6 Hypothesis testing
This chapter is primarily based on Field, A., Miles J., & Field, Z. (2012): Discovering Statistics Using R. Sage Publications, chapters 5, 9, 15, 18.
6.1 Introduction
6.1.1 Why do we test hypotheses?
We test hypotheses because we are confined to taking samples – we rarely work with the entire population. A sample that we take is likely to be different from a second sample that we take from the same population, so we use hypothesis tests to generalize from the sample to the population. However, we need some measure of uncertainty we can assign to analytic results. We use the standard error (i.e., the standard deviation of a large number of hypothetical samples) to gauge how well a particular sample represents the population. This is shown in the following figure.
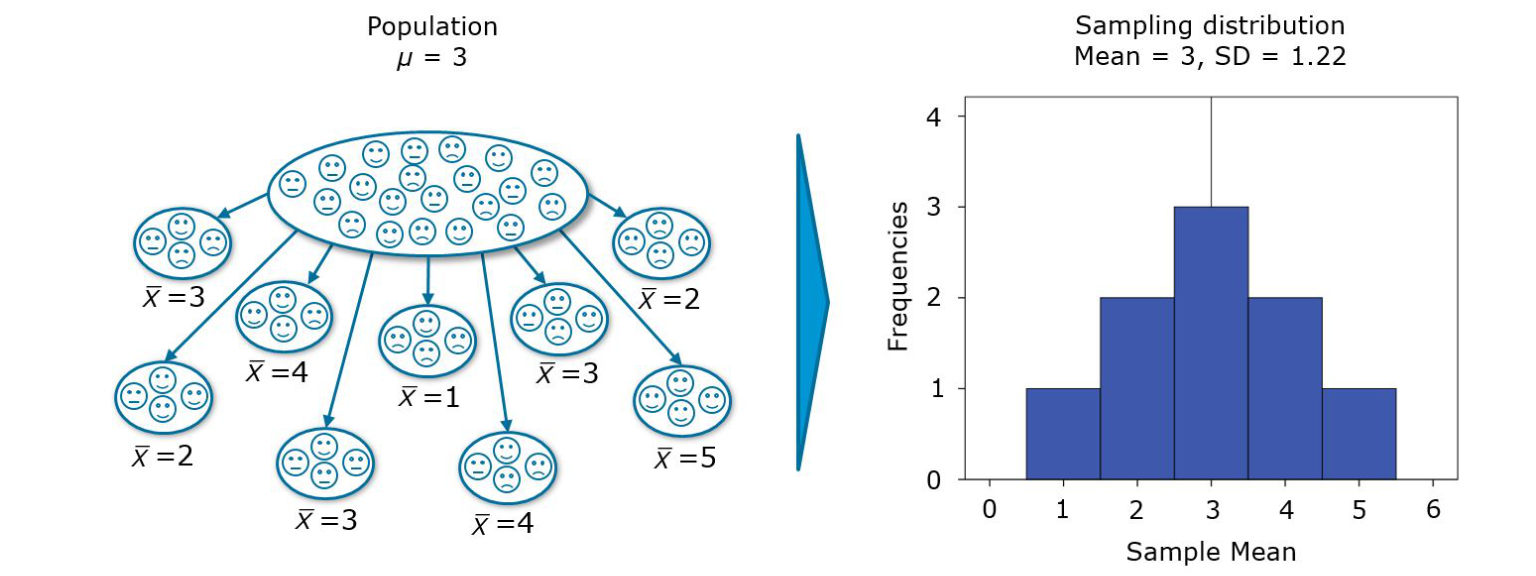
Figure 6.1: Sampling distribution (source: Field, A. et al. (2012): Discovering Statistics Using R, p. 44)
Recall the definition of the standard error (SE):
\[\begin{equation} \begin{split} SE = \frac{s}{\sqrt{n}} \end{split} \tag{6.1} \end{equation}\]where s is the standard deviation and n is the number of observations in our sample. If the standard error is low, then we expect most samples to have similar means. If the standard error is large, large differences in sample means are more likely. If the difference in sample means is large, then we would expect based on the standard error that either the collected samples may be atypical of the population, or that the samples come from different populations but are typical of their respective population.
Note that there is a relationship between the sample size and the standard error. The larger the sample size, the smaller the standard error. This can be attributed to the fact that with larger samples, there is less uncertainty that the sample is a good approximation of the entire population.
The following plot shows the relationship between the sample size and the standard error. A hypothetical population of 1,000,000 observations is simulated and samples of ascending size are randomly drawn from this population. You can see that the standard error is decreasing with the number of observations.
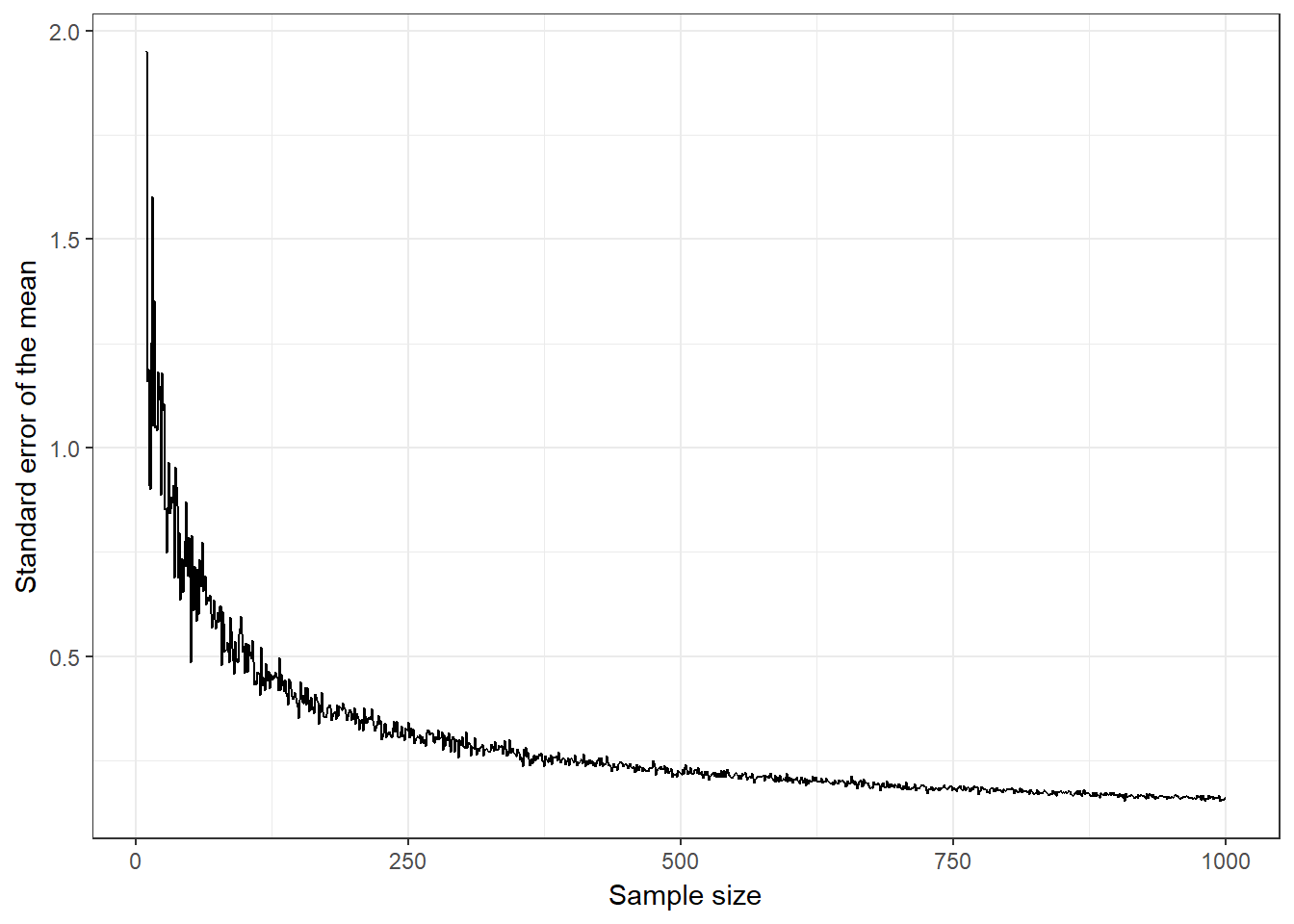
Figure 6.2: Relationship between the sample size and the standard error
Why is this important? Well, the generalized equation for calculating a test statistic is given by
\[\begin{equation} \begin{split} test\ statistic = \frac{effect}{error} \end{split} \tag{6.2} \end{equation}\]where effect denotes the effect that you are investigating (e.g., the mean difference in an outcome variable between two experimental groups), while error denotes the variation we would naturally expect to find based on the variablity in the data and the sample size (e.g., the standard error of the mean difference). When the standard error is lower, the test statistic gets larger and you are more likely to obtain a significant test result. Hence - all else being equal - when you increase your sample size, this will increase the value of your test statistic (i.e., decrease the p-value).
6.1.2 The process of hypothesis testing
The process of hypothesis testing consists of 6 consecutive steps:
- Formulate null and alternative hypotheses
- Select an appropriate test
- Choose the level of significance (α)
- Collect data and calculate the test statistic (TCAL)
- Reject or do not reject H0
- Report results and draw a marketing conclusion
In the following, we will use an example to go through the individual steps.
6.2 Parametric tests
The first basic differentiation we can make is between parametric and non-parametric tests.
Parametric tests require that variables are measured on an interval or ratio scale and that the data follows a known distribution. Particularly, tests based on the normal distribution (e.g., the t-test) require four basic assumptions:
- A normally distributed sampling distribution
- Interval or ratio data
And in addition for independent tests:
- Scores in different conditions are independent (because they come from different people)
- Homogeneity of variance
Non-Parametric tests on the other hand do not require the sampling distribution to be normally distributed (a.k.a. “assumption free tests”). These tests may be used when the variable of interest is measured on an ordinal scale or when the parametric assumptions do not hold. They often rely on ranking the data instead of analyzing the actual scores. By ranking the data, information on the magnitude of differences is lost. Thus, parametric tests are more powerful if the sampling distribution is normally distributed.
6.2.1 Independent-means t-test
As an example of a parametric test, we will discuss the independent- and dependent-means t-test. The independent-means t-test is used when there are two experimental conditions and different units (e.g., participants, products) were assinged to each condition, while the dependent-means t-test is used when there are two experimental conditions and the same units (e.g., participants, products) were observed in both experimental conditions.
Example case: What is the effect of price promotions on the sales of music albums?
Experimental setup: As a marketing manager of a music download store, you are interested in the effect of price on demand. In the music download store, new releases were randomly assigned to an experimental group and sold at a reduced price (i.e., 7.95€), or a control group and sold at the standard price (9.95€). A representative sample of 102 new releases were sampled and these albums were randomly assinged to the experimental groups (i.e., 51 albums per group). The sales were tracked over one day.
Let’s load and investigate the data first:
library(dplyr)
library(psych)
library(ggplot2)
library(Hmisc)
rm(music_sales)
music_sales <- read.table("https://raw.githubusercontent.com/IMSMWU/Teaching/master/MRDA2017/music_experiment.dat",
sep = "\t", header = TRUE) #read in data
music_sales$group <- factor(music_sales$group, levels = c(1:2),
labels = c("low_price", "high_price")) #convert grouping variable to factor
str(music_sales) #inspect data## 'data.frame': 102 obs. of 3 variables:
## $ product_id: int 1 2 3 4 5 6 7 8 9 10 ...
## $ unit_sales: int 6 27 30 24 21 11 18 15 18 13 ...
## $ group : Factor w/ 2 levels "low_price","high_price": 1 1 1 1 1 1 1 1 1 1 ...head(music_sales) #inspect dataInspect frequencies.
table(music_sales$unit_sales, music_sales$group) #frequencies##
## low_price high_price
## 0 0 7
## 1 0 1
## 2 2 4
## 3 8 16
## 4 4 0
## 5 2 0
## 6 18 9
## 9 3 6
## 10 1 0
## 11 1 0
## 12 3 3
## 13 1 0
## 15 2 2
## 18 2 1
## 20 0 1
## 21 1 0
## 24 1 0
## 27 1 0
## 30 1 1Inspect descriptives (overall and by group).
psych::describe(music_sales$unit_sales) #overall descriptives## vars n mean sd median trimmed mad min max range skew kurtosis se
## X1 1 102 7.12 6.26 6 6.1 4.45 0 30 30 1.71 3.02 0.62describeBy(music_sales$unit_sales, music_sales$group) #descriptives by group##
## Descriptive statistics by group
## group: low_price
## vars n mean sd median trimmed mad min max range skew kurtosis se
## X1 1 51 8.37 6.44 6 7.17 4.45 2 30 28 1.66 2.22 0.9
## --------------------------------------------------------
## group: high_price
## vars n mean sd median trimmed mad min max range skew kurtosis se
## X1 1 51 5.86 5.87 3 4.9 4.45 0 30 30 1.84 4.1 0.82Create boxplot and plot of means.
ggplot(music_sales, aes(x = group, y = unit_sales)) +
geom_boxplot() +
labs(x = "Experimental group", y = "Number of sales") +
theme_bw() 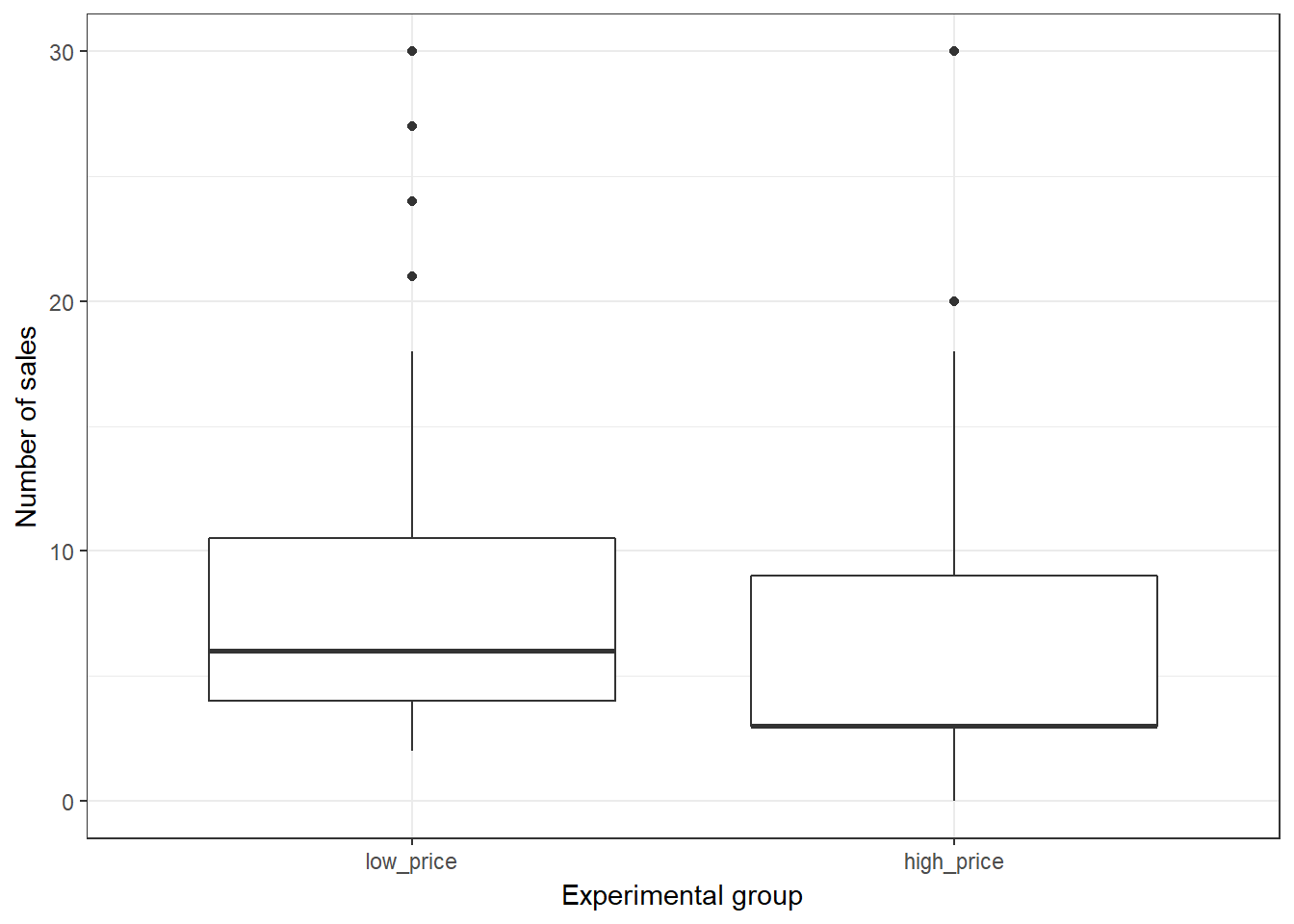
Figure 6.3: Boxplot
ggplot(music_sales, aes(group, unit_sales)) +
geom_bar(stat = "summary", color = "black", fill = "white", width = 0.7) +
geom_pointrange(stat = "summary") +
labs(x = "Group", y = "Average number of sales") +
theme_bw()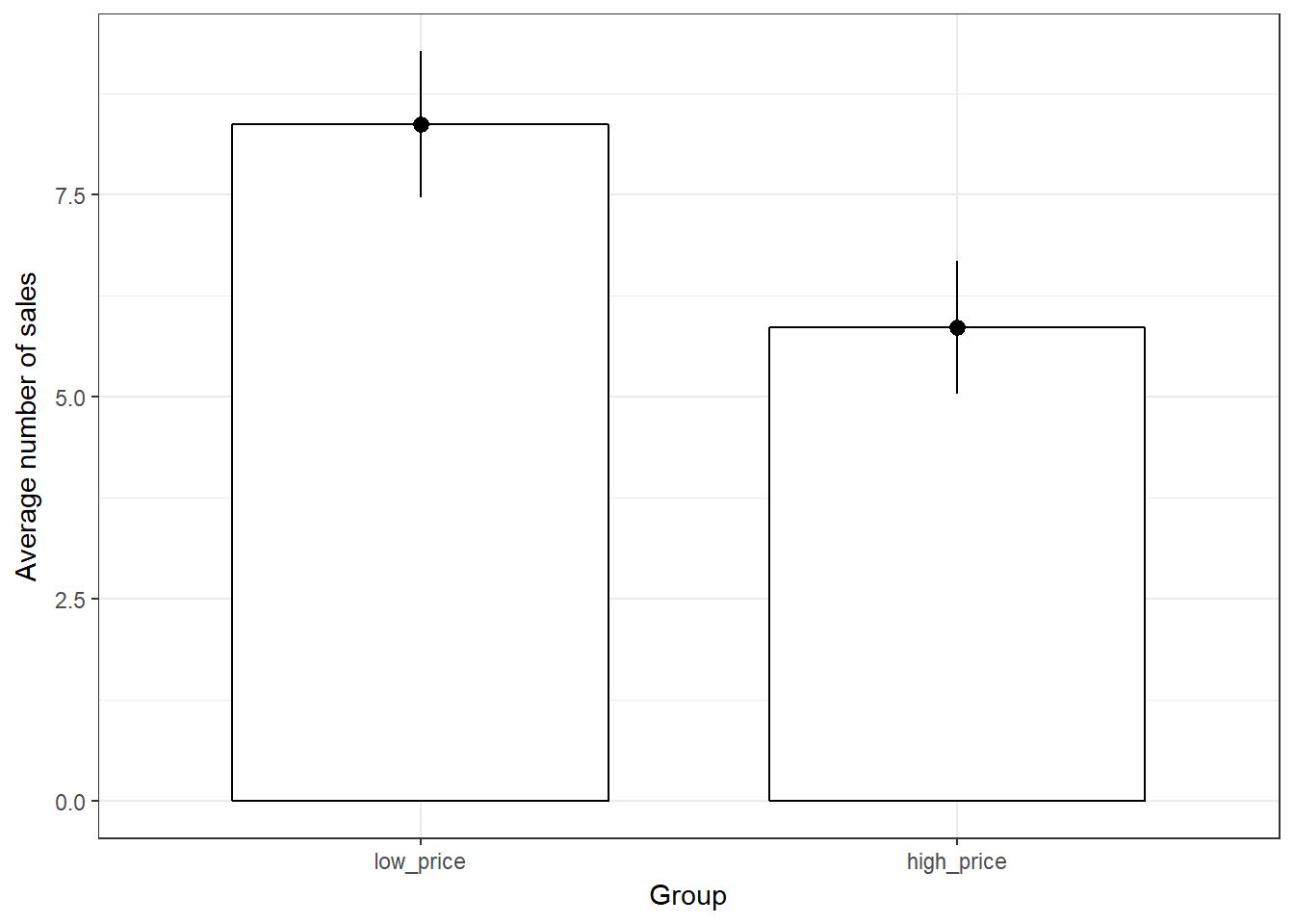
Figure 6.3: Plot of means
ggplot(music_sales,aes(unit_sales)) +
geom_histogram(binwidth = 4, col = "black", fill = "darkblue") +
facet_wrap(~group) +
labs(x = "Number of sales", y = "Frequency") +
theme_bw()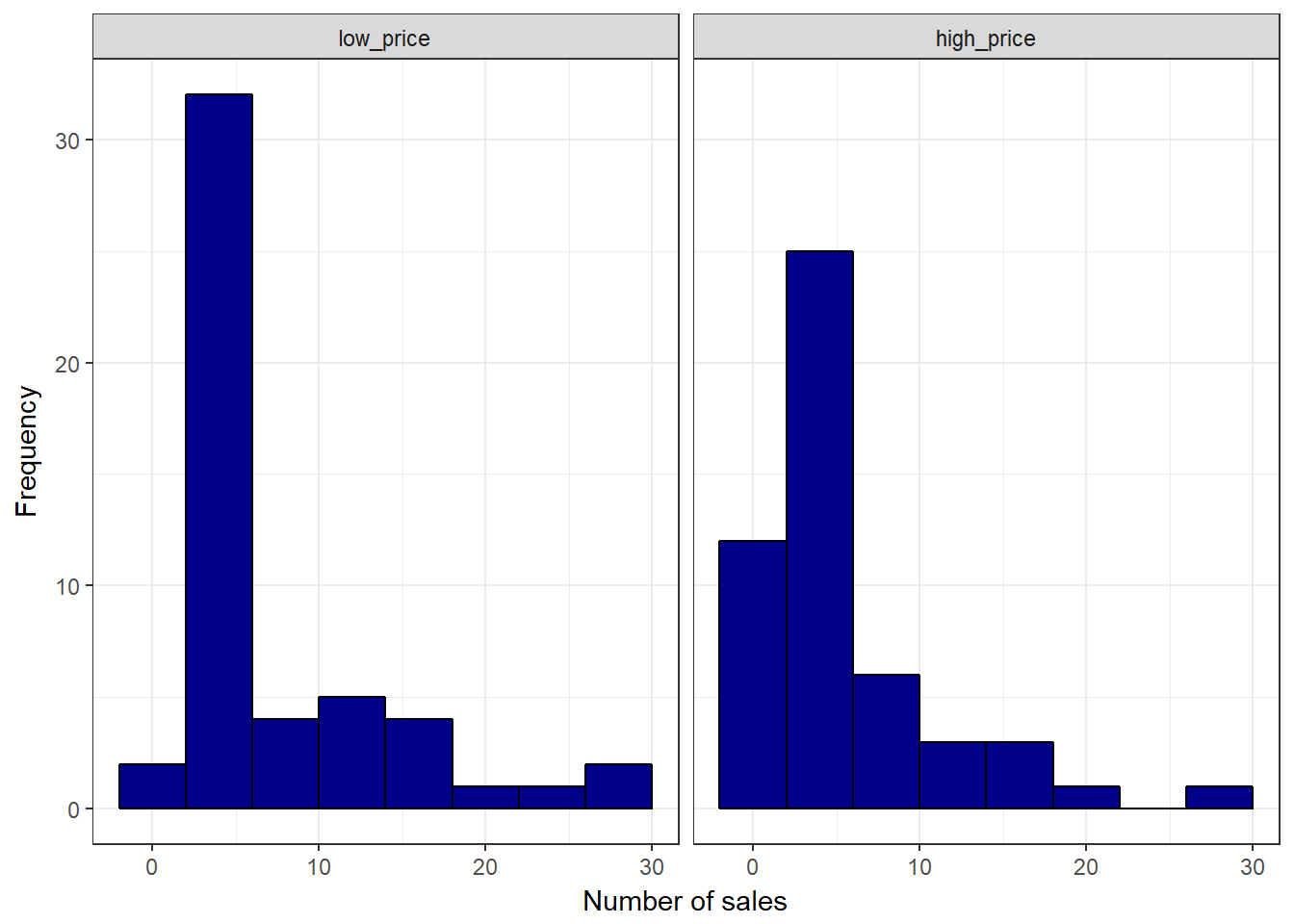
Figure 6.3: Histogram
There appears to be a difference between the two groups. The quesition is: is this difference statstically significant, or would we expect to observe a difference of this magnitude by chance given the sample size and variability in the data?
Let’s test this using a statistical test!
6.2.1.1 Formulate null and alternative hypothesis
What do we mean by null and alternative hypothesis?
Null Hypothesis (H0):
- Is a statement of the status quo, one of no difference or no effect.
- If the null hypothesis is not rejected, no changes will be made.
- Refers to a specified value of the population parameter (e.g., μ), not a sample statistic, (e.g., \(\large\overline{x}\))
- The null hypothesis may be rejected, but it can never be accepted based on a single test. In classical hypothesis testing, there is no way to determine whether the null hypothesis is true.
Alternative Hypothesis (H1):
- Expects some difference or effect.
- Accepting the alternative hypothesis will lead to changes in opinions or actions.
- Rejection of null hypothesis is taken as support for the experimental / alternative hypothesis.
Furthermore, we can differentiate between directional and non-directional hypotheses:
Directional hypothesis:
- States the direction of the effect.
- Leads to a one-tailed test:
\(H_0: \mu_1 \le \mu_2\)
\(H_1: \mu_1 > \mu_2\)
Example: the mean sales of albums sold at 7.95€ are higher than the mean sales of albums sold at 9.95€.
Non-directional hypothesis:
- States that an effect will occur, but it does not state the direction of the effect.
- Leads to a two-tailed test:
\(H_0: \mu_1 = \mu_2\)
\(H_1: \mu_1 \neq \mu_2\)
Example: there is no difference between the mean sales for the two different download prices.
In our example, a non-directional hypothesis would be appropriate. The reason is that we cannot be 100% certain about the direction that the effect might take. For example, although it might seem plausible that lower prices might lead to higher sales, it is also possible that price is perceived as a quality signal, making a product less attractive for certain consumer segments.
⚡ Rejecting the null hypothesis does not prove the alternative hypothesis (we can merely provide support for it). Rather, think of it as the chance of obtaining the data we’ve collected assuming that the null hypothesis is true.
6.2.1.2 Select an appropriate test
We fit a statistical model to the data in order to make predictions about the real world phenomenon. The test statistic measures how close the sample is to the null hypothesis. The test statistic often follows a well-known distribution (e.g., normal, t, or chi-square). To select the correct test, various factors need to be taken into consideration. Some examples are:
- On what scale are your variables measured (categorical vs. continuous)?
- Do you want to test for relationships or differences?
- If you test for differences, how many groups would you like to test?
- For parametric tests, are the assumptions fulfilled?
The following flow chart provides a rough guideline on selecting the correct test:
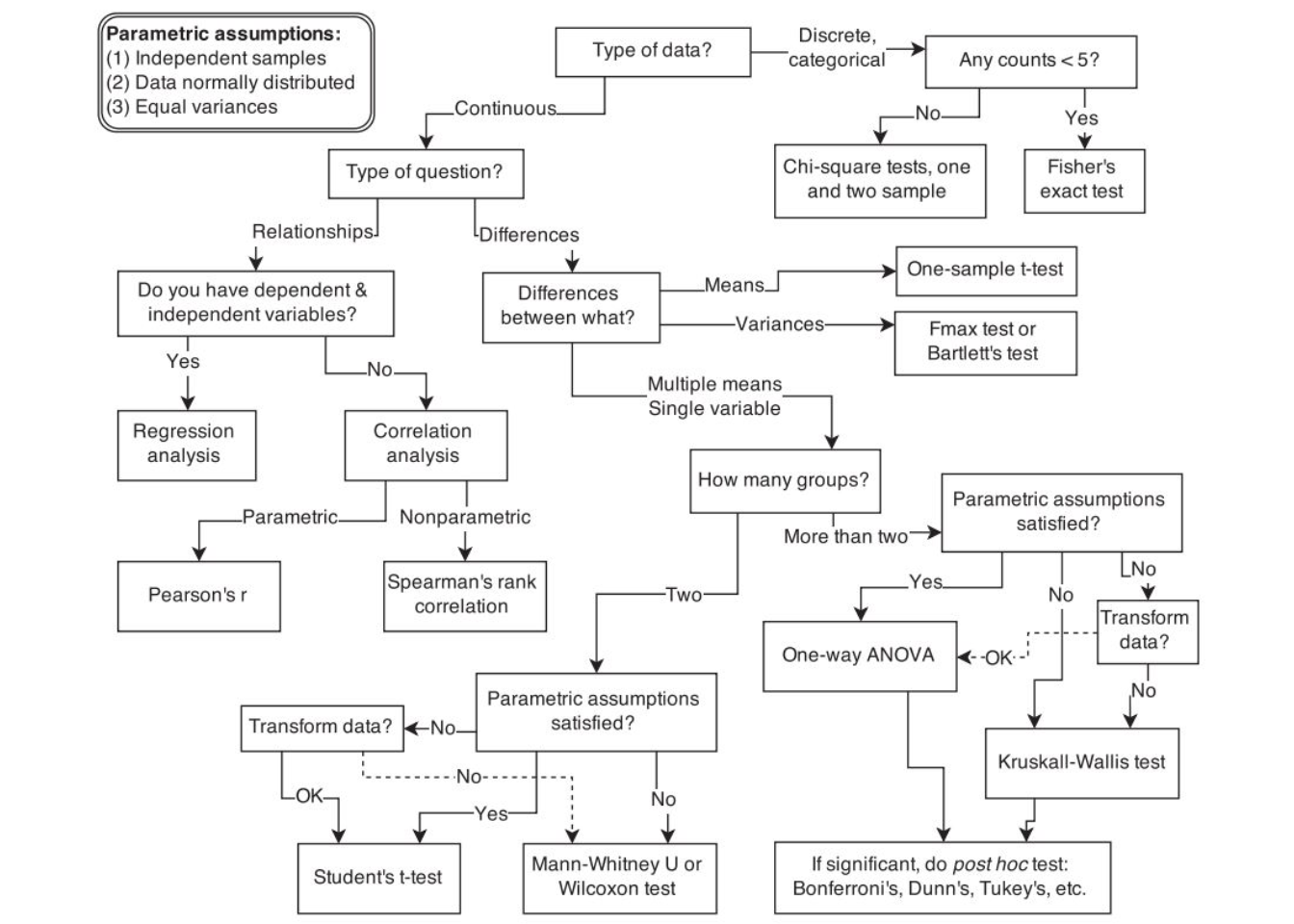
Figure 6.4: Flowchart for selecting an appropriate test (source: McElreath, R. (2016): Statistical Rethinking, p. 2)
For a detailed overview of the different type of tests, please also refer to this overview by the UCLA.
In the present example, the t-test is appropriate since we would like to test for differences between two means of a single continuous variable (sales).
Properties of the t-distribution:
- Adequate when the mean for a normally distributed variable is estimated (Student’s t-distribution).
- Assumes that the mean is known and that the population variance is estimated from a normally distributed sample.
- The t-distribution is similar to the normal distribution in appearance (bell-shaped and symmetric).
- As the number of degrees of freedom increases, the t-distribution approaches the normal distribution.
To see this, the following graph shows the t-distribution with different degrees of freedom for a two-tailed test and α = 0.05.
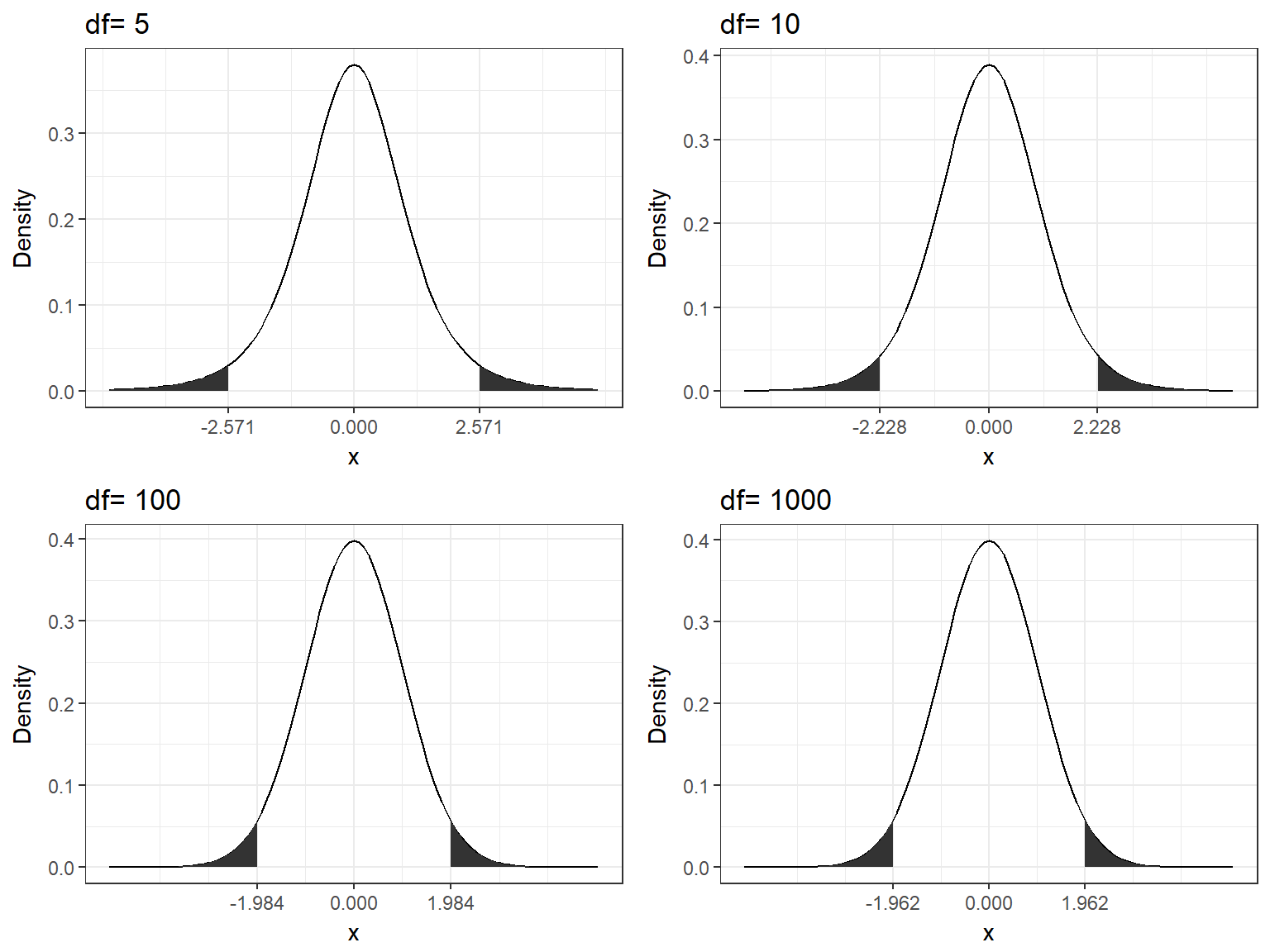
Figure 6.5: Critical values for t-distribution (two-tailed, alpha=0.05)
You can see that the critical value (TCR) decreases as the degrees of freedom increase. This is due to the fact that larger degrees of freedom mean that you have more information available relative to the number of parameters that you would like to estimate.
Connected to the decision on how to phrase the hypotheses (directional vs. non-directional) is the choice of a one-tailed versus a two-tailed test. Let’s first think about the meaning of a one-tailed test. Using a significance level of 0.05, a one-tailed test means that 5% of the total area under the probability distribution of our test statistic is located in one tail. Thus, under a one-tailed test, we test for the possibility of the relationship in one direction only, disregarding the possibility of a relationship in the other direction. In our example, a one-tailed test will test either if the mean sales level in the experimental condition is significantly larger or smaller compared to the control condition, but not both. Depending on the direction, the mean sales level of the experimental group is significantly larger (smaller) if the test statistic is located in the top (bottom) 5% of its probability distribution, resulting in a p-value smaller than 0.05.
The following graph shows the critical values that our test statistic would need to surpass so that the mean difference between the groups would be deemed statistically significant. It can be seen that under a one-sided test, the rejection region is at one end of the distribution or the other. In a two-sided test, the rejection region is split between the two tails. As a consequence, the critical value of the test statistic is smaller using a one-tailed test, meaning that it has more power to detect an effect.
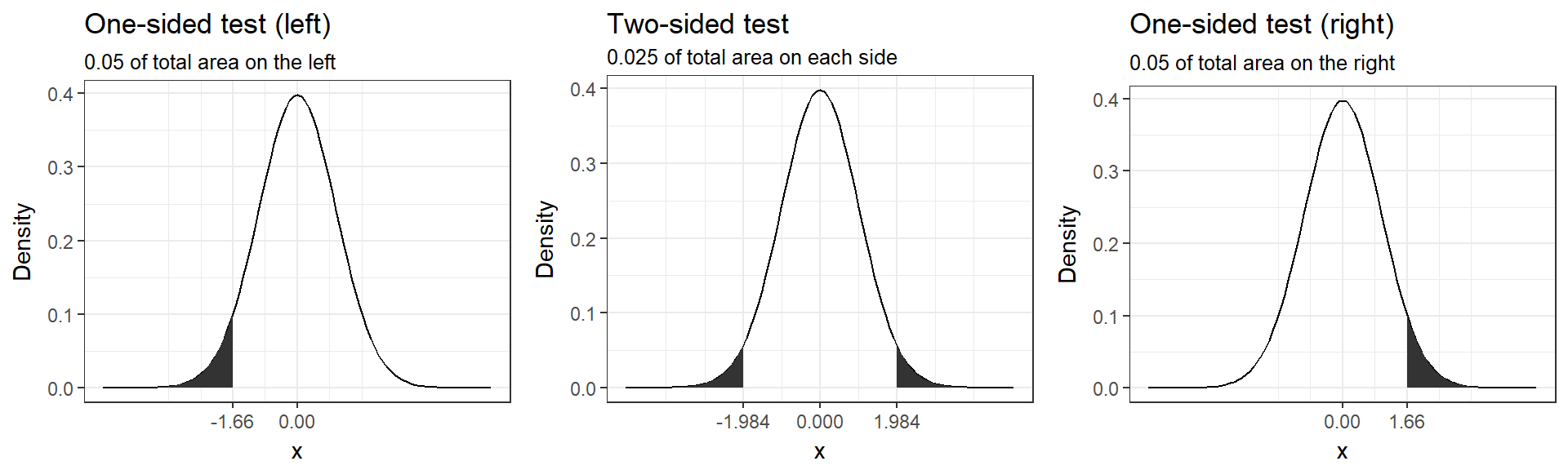
Figure 6.6: One-sided versus two-sided test (df=100, alpha=0.05)
In our example, we used a non-directional hypothesis, meaning that a two-sided test is appropriate. Nevertheless, let’s consider the consequences of using a directional hypothesis (one-sided test). If we assume a positive effect of price promotion on sales (reducing the price leads to higher sales), then we would expect the mean sales in the experimental group to be larger compared to the control group and, thus, derive a positive test statistic. Imagine a scenario where we need a test statistic bigger than 1.66 to find a significant effect at the 5% level under a directional hypothesis, but the test-statistic we get is actually -2.1. In this case, we would retain the null hypothesis although an effect exists and could have been detected using a two-sided test. So unless you consider the consequences of missing an effect in the untested direction negligible, you should use a two-sided test (non-directional hypothesis).
Now we need to make a decision between the independent and the dependent t-test. As stated above, the independent-means t-test is used when there are two experimental conditions and different units were assinged to each condition, while the dependent-means t-test is used when there are two experimental conditions and the same units were observed in both conditions of the experiment. In our example, the independent-means t-test is appropriate since different music albums were randomly assigned to both experimental conditions.
Finally, let’s test if the assumptions of the chosen test are fulfilled:
- A normally distributed sampling distribution (✔ - we can assume this since n > 30 per group)
- Interval or ratio data (✔ - sales is on a ratio scale)
- Scores in different conditions are independent (✔ - products were randomly assigned)
- Homogeneity of variance: you don’t have to worry about this assumption too much for three reasons: 1) violating this assumption only really matters if you have unequal group sizes - in our context, we have equal group sizes (n = 51 per group); 2) even if this assumption is violated, there is an adjustment (called Welch’s t-test) which corrects for unequal variances and this is easily implemented in R (actually, it is the default setting); 3) the tests for homogeneity of variances tend to work better when group sizes are equal and samples are large (which is when we don’t need it), and not so well with unequal group sizes and smaller samples (which is when you actually need it).
6.2.1.3 Choose the level of significance
Next, we need to choose the level of significance (α). It is important to note that the choice of the significance level affects the type 1 and type 2 error:
- Type I error: When we believe there is a genuine effect in our population, when in fact there isn’t. Probability of type I error (α) = level of significance.
- Type II error: When we believe that there is no effect in the population, when in fact there is.
This following table shows the possible outcomes of a test (retain vs. reject H0), depending on whether H0 is true or false in reality.
| Retain H0 | Reject H0 | |
|---|---|---|
| H0 is true | Correct decision: 1-α (probability of correct retention); |
Type 1 error: α (level of significance) |
| H0 is false | Type 2 error: β (type 2 error rate) |
Correct decision: 1-β (power of the test) |
When you plan to conduct an experiment, there are some factors that are under direct control of the researcher:
- Significance level (α): The probability of finding an effect that does not genuinely exist.
- Sample size (n): The number of observations in each group of the experimental design.
Unlike α and n, which are specified by the researcher, the magnitude of β depends on the actual value of the population parameter. In addition, β is influenced by the effect size (e.g., Cohen’s d), which can be used to determine a standardized measure of the magnitude of an observed effect. The following parameters are affected more indirectly:
- Power (1-β): The probability of finding an effect that does genuinely exists.
- Effect size (d): Standardized measure of the effect size under the alternate hypothesis.
Although β is unknown, it is related to α. For example, if we would like to be absolutely sure that we do not falsely identify an effect which does not exist (i.e., make a type I error), this means that the probability of identifying an effect that does exist (i.e., 1-β) decreases and vice verca. Thus, an extremely low value of α (e.g., α = 0.0001) will result in intolerably high β errors. A common approach is to set α=0.05 and β=0.80.
Unlike the t-value of our test, the effect size (d) is unaffected by the sample size and can be categorized as follows (see Cohen, J. 1988):
- 0.2 (small effect)
- 0.5 (medium effect)
- 0.8 (large effect)
In order to test more subtle effects (smaller effect sizes), you need a larger sample size compared to the test of more obvious effects. In this paper, you can find a list of examples for different effect sizes and the number of obervations you need to reliably find an effect of that magnitude. Although the exact effect size is unknown before the experiment, you might be able to make a guess about the effect size (e.g., based on previous studies).
When constructing an experimental design, your goal should be to maximize the power of the test while maintaining an acceptable significance level and keeping the sample as small as possible. To achieve this goal, you may use the pwr package, which let’s you compute n, d, alpha, and power. You only need to specify three of the four input variables to get the fourth.
For example, what sample size do we need (per group) to identify an effect with d = 0.6, α = 0.05, and power = 0.8:
library(pwr)
pwr.t.test(d = 0.6, sig.level = 0.05, power = 0.8,
type = c("two.sample"), alternative = c("two.sided"))##
## Two-sample t test power calculation
##
## n = 44.58577
## d = 0.6
## sig.level = 0.05
## power = 0.8
## alternative = two.sided
##
## NOTE: n is number in *each* groupOr we could ask, what is the power of our test with 51 observations in each group, d = 0.6, and α = 0.05:
pwr.t.test(n = 51, d = 0.6, sig.level = 0.05, type = c("two.sample"),
alternative = c("two.sided"))##
## Two-sample t test power calculation
##
## n = 51
## d = 0.6
## sig.level = 0.05
## power = 0.850985
## alternative = two.sided
##
## NOTE: n is number in *each* group6.2.1.4 Collect data and calculate the test statistic
Step 1: Compute means and variances of samples
The two populations are sampled and the means and variances are computed based on samples of sizes n1 and n2.
Mean of sample 1: \[\begin{equation} \begin{split} \overline{X}_1=\frac{\sum_{i=1}^{n_1} X_i}{n_1} = \frac{427}{51} = 8.37 \end{split} \tag{6.3} \end{equation}\] Variance of sample 1: \[\begin{equation} \begin{split} s_1=\frac{\sum_{i=1}^{n_1} (X_i-\overline{X}_1)^2}{n_1-1}=41.52 \end{split} \tag{6.4} \end{equation}\] Mean of sample 2: \[\begin{equation} \begin{split} \overline{X}_2=\frac{\sum_{i=1}^{n_2} X_i}{n_2} = \frac{564}{12} = 5.86 \end{split} \tag{6.5} \end{equation}\] Variance of sample 2: \[\begin{equation} \begin{split} s_2=\frac{\sum_{i=1}^{n_2} (X_i-\overline{X}_2)^2}{n_2-1}=34.48 \end{split} \tag{6.6} \end{equation}\]
Let’s test if the calculated values are correct:
mean_1 <- mean(music_sales[music_sales$group == "low_price",
"unit_sales"])
mean_1## [1] 8.372549var_1 <- var(music_sales[music_sales$group == "low_price",
"unit_sales"])
var_1## [1] 41.51843mean_2 <- mean(music_sales[music_sales$group == "high_price",
"unit_sales"])
mean_2## [1] 5.862745var_2 <- var(music_sales[music_sales$group == "high_price",
"unit_sales"])
var_2## [1] 34.48078Step 2: Compute pooled variance estimate from the sample variances
When we compare two groups that contain different numbers of participants and the the populations can be expecte to have approximately the same variances, we apply a weighting of the variances of each sample to compute the pooled variance.
\[\begin{equation} \begin{split} s_p=\frac{\sum_{i=1}^{n_1}(X_i-\overline{X}_1)^2+\sum_{i=1}^{n_2}(X_i-\overline{X}_2)^2}{n_1+n_2-2}=\frac{(n_1-1)*s_1^2+(n_2-1)*s_2^2}{n_1+n_2-2}=37.99 \end{split} \tag{6.7} \end{equation}\]
Let’s test if the calculated value is correct:
n_1 <- nrow(music_sales[music_sales$group == "low_price",
])
n_1## [1] 51n_2 <- nrow(music_sales[music_sales$group == "high_price",
])
n_2## [1] 51var_pooled <- ((n_1 - 1) * var_1 + (n_2 - 1) * var_2)/(n_1 +
n_2 - 2)
var_pooled## [1] 37.99961Note that in this case, both groups contain the same number of observations. Therefore, the weighting is not really needed. The weighting is only necessary to account for differences in sample size between groups. In our case, the following would yield the same result because the sample sizes are the same.
var_non_pooled <- (var_1 + var_2)/2
var_non_pooled## [1] 37.99961Step 3: Compute the standard error of the differences
According to the variance sum law, to find the variance of the sampling distribution of differences, we merely need to add together the variances of the sampling distributions of the two populations that we are comparing:
\[\begin{equation} \begin{split} s_{\overline{X}_1-\overline{X}_2}^2={\frac{s_1^2}{N_1}+\frac{s_2^2}{N_2}} \end{split} \tag{6.8} \end{equation}\]Now we have the variance of the sampling distribution of differences. To find the standard error, we only need to take the sqaure root of the variance (because the standard error is the standard deviation of the sampling distribution and the standard error is the square root of the variance):
\[\begin{equation} \begin{split} s_{\overline{X}_1-\overline{X}_2}=\sqrt{{\frac{s_1^2}{N_1}+\frac{s_2^2}{N_2}}}=\sqrt{{\frac{37.99}{51}+\frac{37.99}{51}}}=1.22 \end{split} \tag{6.9} \end{equation}\]Let’s check again if the result is corect:
se_x1_x2 <- sqrt((var_pooled/n_1) + (var_pooled/n_2))
se_x1_x2## [1] 1.22073Step 4: Compute the value of the test statistic
The test statistic can now be calculated as:
Note that \((\mu_1-\mu_2)=0\), because \(H_0: \mu_1=\mu_2\)
And again, we check the result:
t_cal <- (mean_1 - mean_2)/se_x1_x2
t_cal## [1] 2.055987Note that you don’t have to compute these statistics manually! Luckily, there is a function for the t-test, which computes steps 1-4 for you in just one single line of code:
t.test(unit_sales ~ group, data = music_sales)##
## Welch Two Sample t-test
##
## data: unit_sales by group
## t = 2.056, df = 99.15, p-value = 0.04241
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## 0.08765687 4.93195097
## sample estimates:
## mean in group low_price mean in group high_price
## 8.372549 5.862745Note that the degrees of freedom are not 100 (as we would expect), but 99.15 instead. This difference is because of the Welch correction, which adjusts the degrees of freedom based on the homogeneity of variance. R implements this correction by default and it is okay for you to report the adjusted results. However, you could also force R to assume equal variances to exactly replicate the results we derived through manual calculations using the var.equal=TRUE argument. In this case you would first need to test if this assumption is fulfilled using Levene’s Test, which tests H0 = group variances are equal; H1 = group variances are not equal (the leveneTest(...) function is in the car package).
library(car)
leveneTest(unit_sales ~ group, data = music_sales)## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 1 0.0085 0.9269
## 100In this case, we fail to reject the Null of equal variances (p > 0.05), hence the variances are equal (however, see above regarding the concerns about this test). Now we run the t-test using the var.equal=TRUE argument:
t.test(unit_sales ~ group, data = music_sales, var.equal = TRUE)##
## Two Sample t-test
##
## data: unit_sales by group
## t = 2.056, df = 100, p-value = 0.04239
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## 0.08791121 4.93169663
## sample estimates:
## mean in group low_price mean in group high_price
## 8.372549 5.862745As you can see, the results are fairly similar and it is fine to rely solely on the corrected results (i.e., Welch’s t-test).
Effect size
If you wish to obtain a standardized measure of the effect, you may compute the effect size (Cohen’s d) as follows:
d <- (mean_1 - mean_2)/sqrt(var_pooled + var_pooled/2)
d## [1] 0.3324334According to the thresholds defined above, this effect would be judged to be a small-medium effect.
6.2.1.5 Reject or do not reject H0
There are different (albeit interrelated) ways of deciding whether a hypothesis should be rejected:
6.2.1.5.1 p-value
- Determine the probability associated with the test statistic TSCAL (p-value).
- Compare the p-value with the level of significance, α.
- The p-value is the likelihood of observing a result when the null hypothesis is true (probability of obtaining a particular value).
- A small p-value signals that it is unlikely to observe TSCAL under the null hypothesis.
- You reject H0 if the p-value is smaller than α.
The p-value is included in the output from the t-test above. However, you can calculate the p-value for TCAL manually as follows:
df <- (n_1 + n_2 - 2)
2 * (1 - pt(abs(t_cal), df))## [1] 0.04238901Here, pt() is the cumulative probability distribution function of the t-distribution. Cumulative probability means that the function returns the probability that the test statistic will take a value less than or equal to the calculated test statistic given the the degrees of freedom. However, we are interested in obtaining the probability of observing a test statistic larger than or equal to the calculated test statstic under the null hypothsis (i.e., the p-value). Thus, we need to subtract the cumulative probability from 1. In addition, since we are runnig a two-sided test, we need to multiply the probability by 2 to account for the rejection region at the other side of the distribution.
Interpretation: Under the assumption that the null hypothesis is true the probability of finding a difference of at least the observed magnitude is less than 5%.
Decision: Reject H0, given that the p-value of 0.042 is smaller than 0.05.
However, remember that the p-value does not tell us anything about the likelihood of the alternative hypothesis!
6.2.1.5.2 t-value
- Determine the critical value of the test statistic (TSCR) in order to define the rejection region and the non-rejection region.
- Determine if TSCAL falls into rejection or non-rejection region.
- If the (absolute) calculated value of the test statistic (TCAL) is greater than the (absolute) critical value of the test statistic (TCR), H0 is rejected.
- If the (absolute) calculated value of the test statistic (TCAL) is larger than the (absolute) critical value of the test statistic (TCR) , the null hypothesis can not be rejected.
However, remember that this does not mean that the null hypothesis is true!
The calculated test statistic is included in the output from the t-test above. In order to determine if the calculated test statistic is larger than the critical value, you can refer to the probability table of the t-distribution to look up the critical value for the given degrees of freedom. However, you can also test if the calculated test statistic is larger than the critical value manually using the qt()-function:
df <- (n_1 + n_2 - 2)
t_cal > qt(0.975, df)## [1] TRUEWe use 0.975 and not 0.95 since we are running a two-sided test and need to account for the rejection region at the other end of the distribution. The output suggests that the calculated t-value is larger than the critical value for the given level of α.
Interpretation: The probability of observing a difference in means of this magnitude or larger is less than 5%.
Decision: Reject H0, given that the calculated t-value (TCAL) of 2.0559868 is larger than critial value (TCR) of 1.9839715.
The following graph shows the test result.
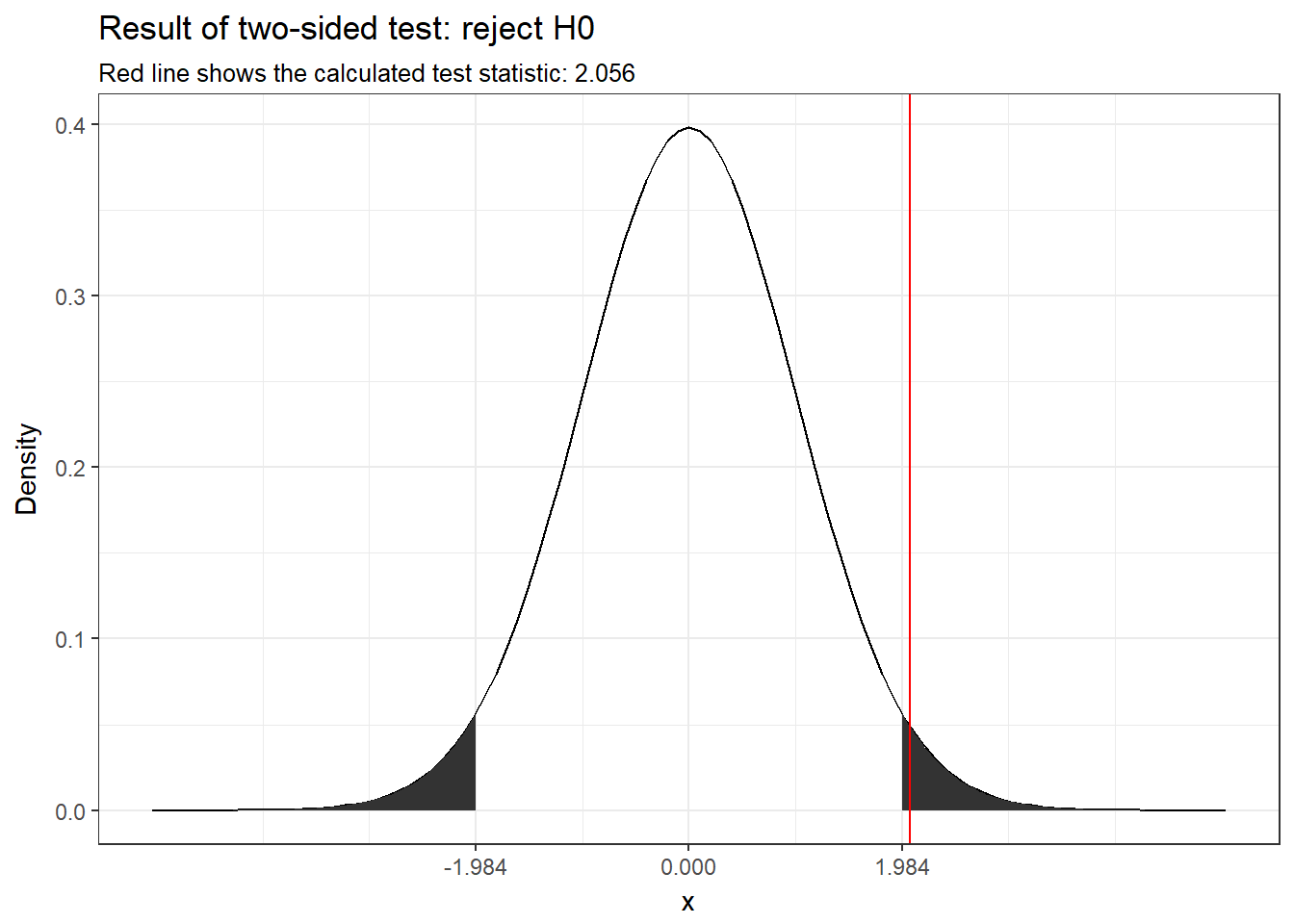
Figure 6.7: Visual depiction of the test result
You can see that the calculated test-statistic (red line) falls into the rejection region.
6.2.1.5.3 Confidence interval
For a given statistic calculated for a sample of observations (e.g., difference in means), the confidence interval is a range around that statistic that is believed to contain, with a certain probability (e.g., 95%), the true value of that statistic under (hypothetical) repeated sampling. In our example, the 95% confidence interval is constructed such that in 95% of samples, the true value of the population mean difference will fall within its limits.
The computation of the 95% confidence interval for the t-test is:
\[\begin{equation} \begin{split} CI = (\overline{X}_1-\overline{X}_2)\pm t_{1-\frac{\alpha}{2}}*\sqrt{{\frac{s_p^2}{N_1}+\frac{s_p^2}{N_2}}} \end{split} \tag{4.1} \end{equation}\]Luckily, the interval is already given in the output of the t-test above. However, we can also compute the interval manually:
(mean_1 - mean_2) - qt(0.975, df) * se_x1_x2## [1] 0.08791121(mean_1 - mean_2) + qt(0.975, df) * se_x1_x2## [1] 4.931697If the parameter value specified in the null hypothesis (usually 0) does not lie within the bounds, we reject H0.
Interpretation: With a 95% probability the mean difference between the populations will lie within [0.09; 4.93].
Decision: Reject H0, given that the true parameter (zero) is not included in the interval.
To illustrate this, we can randomly draw 100 samples from the two populations using simulated data, copute their means and construct confidence intervals around their mean difference. The result is shown in the following graph:
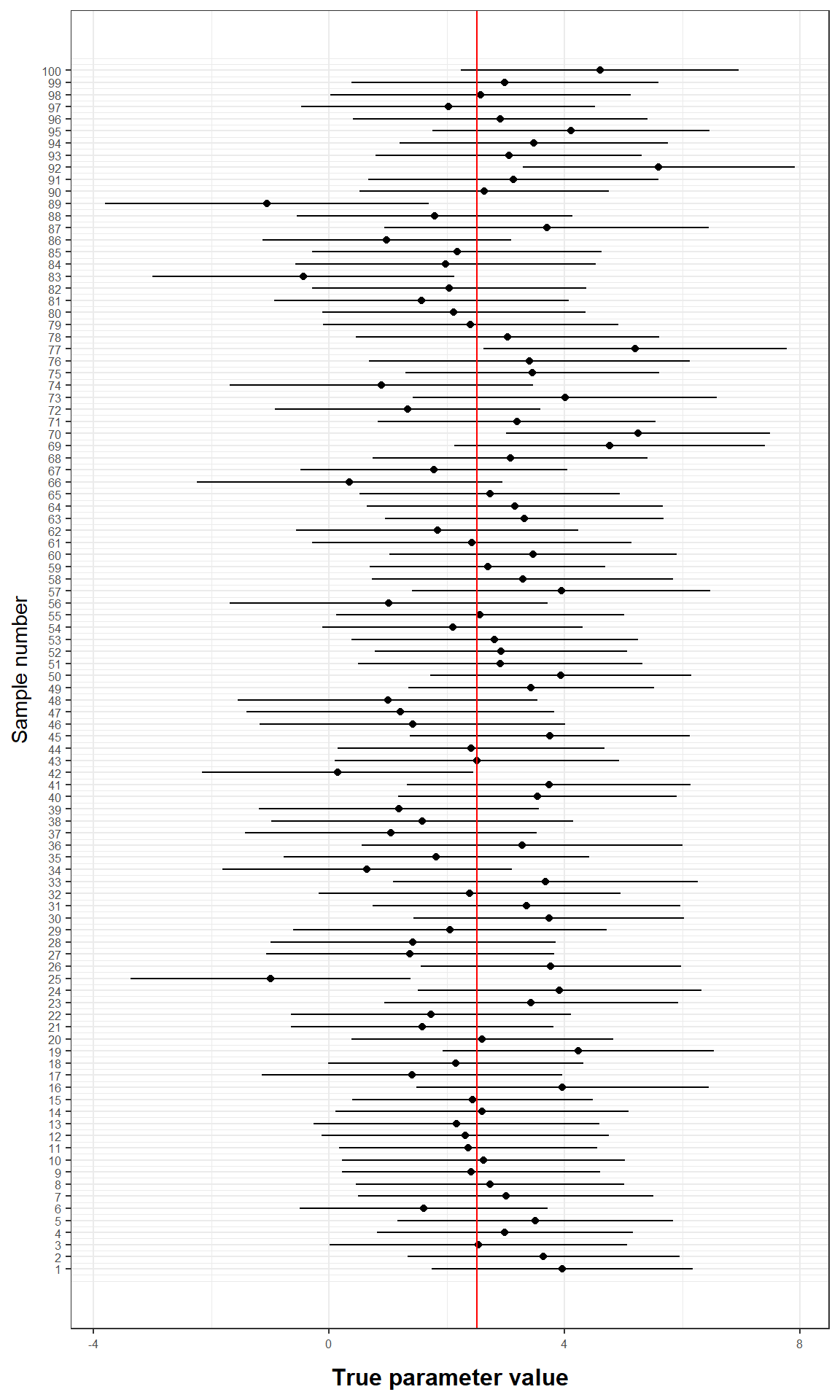
Figure 6.8: Confidence Intervals
You can see that for 5 out of 100 samples, the caclulated confidence interval does not contain the true population mean (red line).
6.2.1.6 Report results and draw a marketing conclusion
The conclusion reached by hypothesis testing must finally be expressed in terms of the marketing research problem:
The test showed that the sales promotion indreased sales by about 2.51 unit on average. This difference is significant t(100) = 2.056, p < .05 (95% CI = [0.09,4.93]).
The following video summarizes how to conduct an independent-means t-test in R
6.2.1.7 A note on the interpretation of p-values
From my experience, students tend to place a lot of weight on p-values when interpreting their research findings. It is therefore important to note some points that hopefully help to put the meaning of a “singnificant” vs. “insignificant” test result into perspective.
Significant result
- Even if the probability of the effect being a chance result is small (e.g., less than .05) it doesn’t necessarily mean that the effect is important.
- Very small and unimportant effects can turn out to be statistically significant if the sample size is large enough.
Insignificant result
- If the probability of the effect occurring by chance is large (greater than .05), the alternative hypothesis is rejected. However, this does not mean that the null hypothesis is true.
- Although an effect might not be large enough to be anything other than a chance finding, it doesn’t mean that the effect is zero.
- In fact, two random samples will always have slightly different means that would deemed to be statistically significant if the samples were large enough.
Thus, you should not base your research conclusion on p-values alone!
It is also crucial to determine the sample size before you run the experiment or before you start your analysis. Why? Consider the following example:
- You run an experiment
- After each respondent you analyze the data and look at the mean difference between the two groups with a t-test
- You stop when you have a significant effect
This is called p-hacking and should be avoided at all costs. Assuming that both groups come from the same population (i.e., there is no difference in the means): What is the likelihood that the result will be significant at some point? In other words, what is the likelihood that you will draw the wrong conclusion from your data that there is an effect, while there is none? This is shown in the following graph using simulated data - the color red indicates significant test results that arise although there is no effect (i.e., false positives).
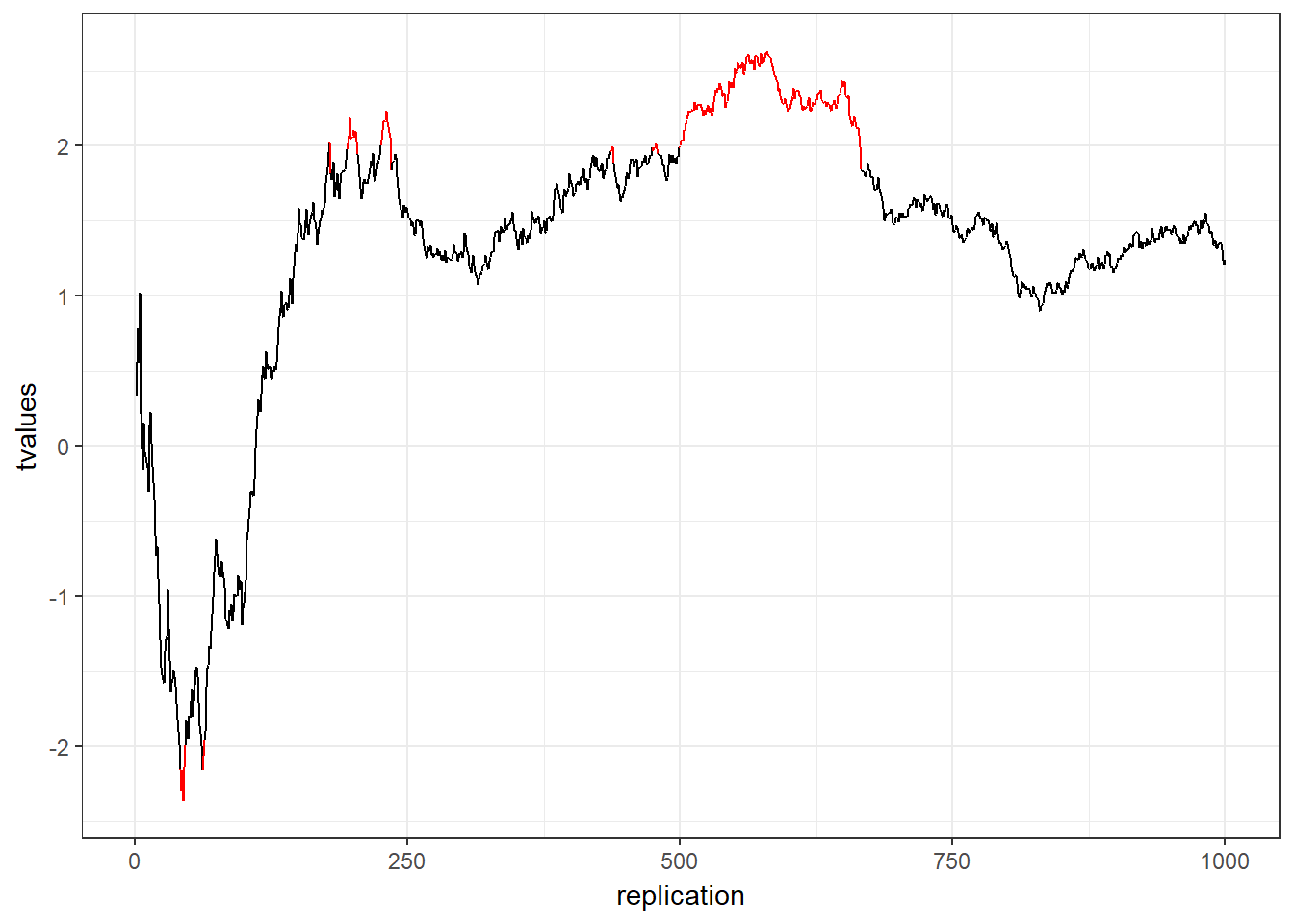
Figure 6.9: p-hacking (red indicates false positives)
6.2.2 Dependent-means t-test
While the independent-means t-test is used when different units (e.g., participants, products) were assinged to the different condition, the dependent-means t-test is used when there are two experimental conditions and the same units (e.g., participants, products) were observed in both experimental conditions.
Imagine, for example, a slightly different experimental setup for the price experiment:
Example case: What is the effect of personalized, dynamic pricing on the sales of music albums?
Experimental setup: In a music download store, new releases were either sold at a reduced price (i.e., 7.95€), or at the standard price (9.95€). Every time a customer came to the store, the prices were randomly determined for every new release. This means that the same 51 albums were either sold at the standard price or at the reduced price and this price was determined randomly. The sales were then recorded over one day. Note the difference to the previous case, where we randomly split the sample and assigned 50% of products to each condition. Now, we randomly vary prices for all albums between high and low prices.
Let’s load and inspect the data:
rm(music_sales_dep)
music_sales_dep <- read.table("https://raw.githubusercontent.com/IMSMWU/Teaching/master/MRDA2017/music_experiment_dependent.dat",
sep = "\t", header = TRUE) #read in data
str(music_sales_dep) #inspect data## 'data.frame': 51 obs. of 3 variables:
## $ product_id : int 1 2 3 4 5 6 7 8 9 10 ...
## $ unit_sales_low_price : int 6 27 30 24 21 11 18 15 18 13 ...
## $ unit_sales_high_price: int 9 12 30 18 20 15 2 3 3 9 ...head(music_sales_dep) #inspect dataThe data are actually the same as before, only now we assume that the observations at different price points come from the same users.
Since we are working with the same data (in a different format), the descriptives are the same as before:
table(music_sales_dep[, c("unit_sales_low_price")]) #frequencies low price##
## 2 3 4 5 6 9 10 11 12 13 15 18 21 24 27 30
## 2 8 4 2 18 3 1 1 3 1 2 2 1 1 1 1table(music_sales_dep[, c("unit_sales_high_price")]) #frequencies high price##
## 0 1 2 3 6 9 12 15 18 20 30
## 7 1 4 16 9 6 3 2 1 1 1psych::describe(music_sales_dep[, c("unit_sales_low_price",
"unit_sales_high_price")]) #overall descriptives## vars n mean sd median trimmed mad min max range
## unit_sales_low_price 1 51 8.37 6.44 6 7.17 4.45 2 30 28
## unit_sales_high_price 2 51 5.86 5.87 3 4.90 4.45 0 30 30
## skew kurtosis se
## unit_sales_low_price 1.66 2.22 0.90
## unit_sales_high_price 1.84 4.10 0.82To plot the data, we need to do some restructuring first, since the variables are now stored in two different columns (“unit_sales_low_price” and “unit_sales_high_price”). This is also known as the “wide” format. To plot the data we need all observations to be stored in one variable. This is also known as the “long” format. We can use the melt(...) function from the reshape2package to “melt” the two variable into one column to plot the data. Since we are working with the same data as before, the plot ist the same as before.
library(reshape2)
ggplot(data = melt(music_sales_dep[, c("unit_sales_low_price",
"unit_sales_high_price")]), aes(x = variable, y = value)) +
geom_bar(stat = "summary", color = "black", fill = "white",
width = 0.7) + geom_pointrange(stat = "summary") +
labs(x = "Group", y = "Average number of sales") +
theme_bw()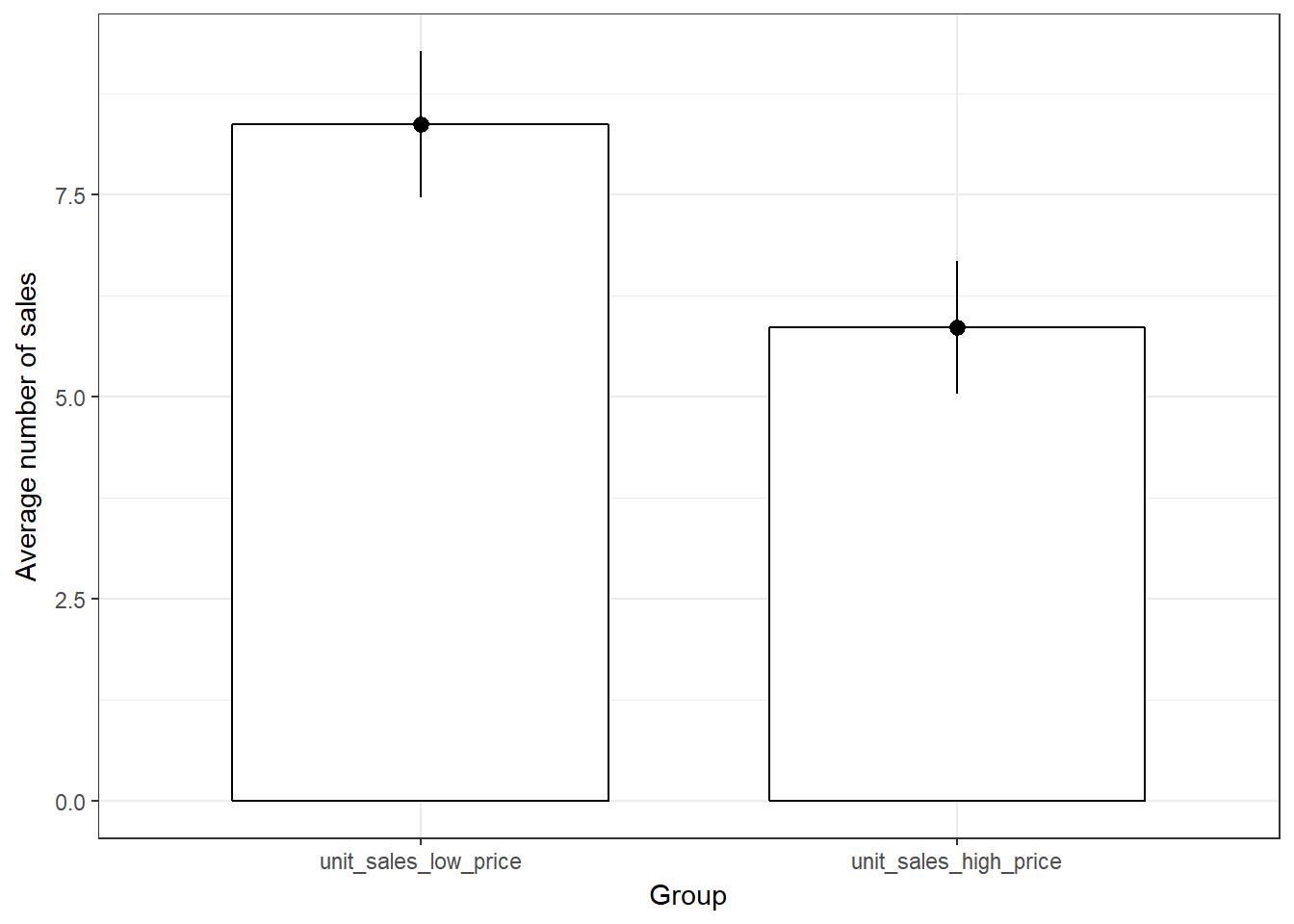
Figure 6.10: plot of means (dependent test)
Suppose we want to test the hypothesis that there is no difference in means between the two groups.
\(H_0: \mu_D = 0\)
\(H_1: \mu_D \neq 0\)
Where D denotes the difference between the observations from the two price groups. Let’s compute a new variable which is the difference between the price groups. H0 states that this difference should be 0.
music_sales_dep$difference <- music_sales_dep$unit_sales_low_price -
music_sales_dep$unit_sales_high_price
music_sales_depThe mean of the paired differences is
\[\begin{equation} \begin{split} \overline{D}=\frac{\sum_{i=1}^{n}D_i}{n}=\frac{128}{51}=2.510 \end{split} \tag{6.11} \end{equation}\]mean_d <- sum(music_sales_dep$difference)/length(music_sales_dep$difference)
mean_d## [1] 2.509804The standard deviation of the paired differences is
\[\begin{equation} \begin{split} s_D = \sqrt\frac{\sum_{i=1}^{n}(D_i-\overline{D})^2}{n-1}=5.651 \end{split} \tag{6.12} \end{equation}\]and the standard error of the paired differences is
\[\begin{equation} \begin{split} s_\overline{D} = \frac{s_D}{\sqrt{n}}=\frac{5.651}{\sqrt{51}}=0.791 \end{split} \tag{6.13} \end{equation}\]sd_d <- sd(music_sales_dep$difference) #standard deviation
sd_d## [1] 5.651097n_d <- nrow(music_sales_dep) #number of observations
n_d## [1] 51se_d <- sd(music_sales_dep$difference)/sqrt(n_d) #standard error
se_d## [1] 0.7913119The test statistic is therefore:
\[\begin{equation} \begin{split} t = \frac{\overline{D}-\mu_d}{s_\overline{D}}=\frac{2.510-0}{0.791}=3.172 \end{split} \tag{6.14} \end{equation}\]on 50 (i.e., n-1) degrees of freedom
t_cal <- mean_d/se_d
t_cal## [1] 3.1717Note that although we used the same data as before, the calculated test statistic is larger compared to the independent t-test. This is because in the dependent sample test, the observations come from the same observational units (i.e., products). Hence, there is no unsystematic variation due to potential differences between products that were assigned to the experimental groups. This means that the influence of unobserved factors (unsystematic variation) relative to the variation due to the experimental manipulation (systematic variation) is not as strong in the dependent-means test compared to the independent-means test.
The confidence interval can be computed as follows:
df <- nrow(music_sales_dep) - 1
(mean_d) - qt(0.975, df) * se_d## [1] 0.9204072(mean_d) + qt(0.975, df) * se_d## [1] 4.099201Again, we don’t have to compute all this by hand since the t.test(...) function can be used to do it for us. Now we have to use the argument paired=TRUE to let R know that we are working with dependent observations.
t.test(music_sales_dep$unit_sales_low_price, music_sales_dep$unit_sales_high_price,
paired = TRUE)##
## Paired t-test
##
## data: music_sales_dep$unit_sales_low_price and music_sales_dep$unit_sales_high_price
## t = 3.1717, df = 50, p-value = 0.00259
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## 0.9204072 4.0992007
## sample estimates:
## mean of the differences
## 2.509804The p-value associated with the test statistic is already reported in the output. However, we could still validate this result manually to see if the calculated test statistic is larger than the critical value:
df <- (n_d - 1) #degrees of freedom
qt(0.975, df) #critical value## [1] 2.008559t_cal #calculated value## [1] 3.1717t_cal > qt(0.975, df) #check if calculated value is larger than critical## [1] TRUEFinally, we need to report the results:
On average, the same music albums sold more units at the lower price point (M = 8.37, SE = 0.902) compared to the higher price point (M = 5.86, SE = 0.822). This difference was significant t(50) = 3.172, p < .01 (95% CI = [0.920, 4.099]).
The following video summarizes how to conduct an dependent-means t-test in R
6.2.3 One-sample t-test
If we are not interested in comparing groups, but would like to test if the mean of a variable is different from a specified threshold, we can use the one-sample t-test. Suppose we wanted to test the hypothesis that the overall mean sales in the “music_experiment.dat” exceeds 4.0 at a significance level of α = 0.05. The hypothesis would be:
\(H_0: \mu \le 4.0\)
\(H_1: \mu > 4.0\)
The estimate of the standard error is simply the standard deviation over the square root of n:
\[\begin{equation} \begin{split} s_\overline{x}=\frac{s_x}{\sqrt{n}}=\frac{6.26}{\sqrt{102}}=0.62 \end{split} \tag{6.15} \end{equation}\]and the t-value is simply
\[\begin{equation} \begin{split} t=\frac{\overline{X}-\mu}{s_\overline{x}}=\frac{7.12-4}{0.62}=5.03 \end{split} \tag{6.16} \end{equation}\]The degrees of freedom for the t-statistic to test the hypothesis about one mean are n-1 (= 101). From the t distribution, the probability of getting a more extreme value than 5.03 is less than 0.05. Hence, H0 is rejected. Alternatively, the critical t-value for 101 degrees of freedom and a significance level of 0.05 is 1.645. This implies that the average sales level exceeds 4.0.
This can be implemented using the t.test function and specifying mu as the threshold for your test and using the argument alternative to specify the direcition in which we would like to test.
t.test(music_sales$unit_sales, mu = 4, alternative = "greater")##
## One Sample t-test
##
## data: music_sales$unit_sales
## t = 5.0281, df = 101, p-value = 0.000001076
## alternative hypothesis: true mean is greater than 4
## 95 percent confidence interval:
## 6.088332 Inf
## sample estimates:
## mean of x
## 7.117647Report the results:
On average, the sales level in our sample exceeded the value 4.0 (M = 7.12, SE = 0.620). This difference was significant t(101) = 5.028, p < .05.
6.3 Non-parametric tests
When should you use non-parametric tests?
- When your DV is measured on an ordinal scale.
- When your data is better represented by the median (e.g., there are outliers that you can’t remove).
- When the assumptions of parametric tests are not met (e.g., normally distributed sampling distribution).
- You have a very small sample size (i.e., the central limit theorem does not apply).
6.3.1 Mann-Whitney U Test (a.k.a. Wilcoxon rank-sum test)
The Mann-Whitney U test is a non-parametric test of differences between groups, similar to the two sample t-test. In contrast to the two sample t-test it only requires ordinally scaled data and relies on weaker assumptions. Thus it is often useful if the assumptions of the t-test are violated, especially if the data is not on a ratio scale, the data is not normally distributed or if the variances can not be assumed to be homogenous. The following assumptions must be fulfilled for the test to be applicable:
- The dependent variable is at least ordinally scaled (i.e. a ranking between values can be established).
- The independent variable has only two levels.
- A between-subjects design is used.
- The subjects are not matched across conditions.
Intuitively, the test compares the frequency of low and high ranks between groups. Under the null hypothesis, the amount of high and low ranks should be roughly equal in the two groups. This is achieved through comparing the expected sum of ranks to the actual sum of ranks.
The test is implemented in R as the function wilcox.test() and there is no need to compute the ranks before you rund the test as the function does this for you. Using the same data on music sales as before the test could be executed as follows:
wilcox.test(unit_sales ~ group, data = music_sales) #Mann-Whitney U Test##
## Wilcoxon rank sum test with continuity correction
##
## data: unit_sales by group
## W = 1710, p-value = 0.005374
## alternative hypothesis: true location shift is not equal to 0The p-value is smaller than 0.05, which leads us to reject the null hypothesis, i.e. the test yields evidence that the price promotion lead to higher sales.
6.3.2 Wilcoxon signed-rank test
The Wilcoxon signed-rank test is a non-parametric test used to analyze the difference between paired observations, analogously to the paired t-test. It can be used when measurements come from the same observational units but the distributional assumptions of the paired t-test do not hold, since it does not require any assumptions about the distribution of the measurements. Since we subtract two values, however, the test requires that the dependent variable is at least interval scaled, meaning that intervals have the same meaning for different points on our measurement scale.
Under the null hypothesis, the differences of the measurements should follow a symmetric distribution around 0, meaning that, on average, there is no difference between the two matched samples. H1 states that the distributions mean is non-zero.
The test can be performed with the same command as the Mann-Whitney U test, provided that the argument paired is set to TRUE.
wilcox.test(music_sales_dep$unit_sales_low_price, music_sales_dep$unit_sales_high_price,
paired = TRUE) #Wilcoxon signed-rank test##
## Wilcoxon signed rank test with continuity correction
##
## data: music_sales_dep$unit_sales_low_price and music_sales_dep$unit_sales_high_price
## V = 867.5, p-value = 0.004024
## alternative hypothesis: true location shift is not equal to 0Using the 95% confidence level, the result would suggest a significant effect of price on sales (i.e., p < 0.05).
The following video summarizes how to conduct non-parametric tests in R
6.4 Categorical data
6.4.1 Comparing proportions
[This section is based on E. McDonnell Feit “Business Experiments - Analyzing A/B Tests”]
In some instances, you will be confronted with differences between proportions, rather than differences between means. For example, you may conduct an A/B-Test and wish to compare the conversion rates between two advertising campaigns. In this case, your data is binary (0 = no conversion, 1 = conversion) and the sampling distribution for such data is binomial. While binomial probabilities are difficult to calculate, we can use a Normal approximation to the binomial when n is large (>100) and the true likelihood of a 1 is not too close to 0 or 1.
Let’s use an example: assume a call center where service agents call potential customers to sell a product. We consider two call center agents:
- Service agent 1 talks to 300 customers and gets 200 of them to buy (conversion rate=2/3)
- Service agent 2 talks to 300 customers and gets 100 of them to buy (conversion rate=1/3)
As always, we load the data first:
call_center <- read.table("https://raw.githubusercontent.com/IMSMWU/Teaching/master/MRDA2017/call_center.dat",
sep = "\t", header = TRUE) #read in data
call_center$conversion <- factor(call_center$conversion,
levels = c(0:1), labels = c("no", "yes")) #convert to factor
call_center$agent <- factor(call_center$agent, levels = c(0:1),
labels = c("agent_1", "agent_2")) #convert to factorNext, we create a table to check the relative frequncies:
rel_freq_table <- as.data.frame(prop.table(table(call_center),
2)) #conditional relative frequencies
rel_freq_tableWe could also plot the data to visualize the frequencies using ggplot:
ggplot(rel_freq_table, aes(x = agent, y = Freq, fill = conversion)) + #plot data
geom_col(width = .7) + #position
geom_text(aes(label = paste0(round(Freq*100,0),"%")), position = position_stack(vjust = 0.5), size = 4) + #add percentages
ylab("Proportion of conversions") + xlab("Agent") + # specify axis labels
theme_bw()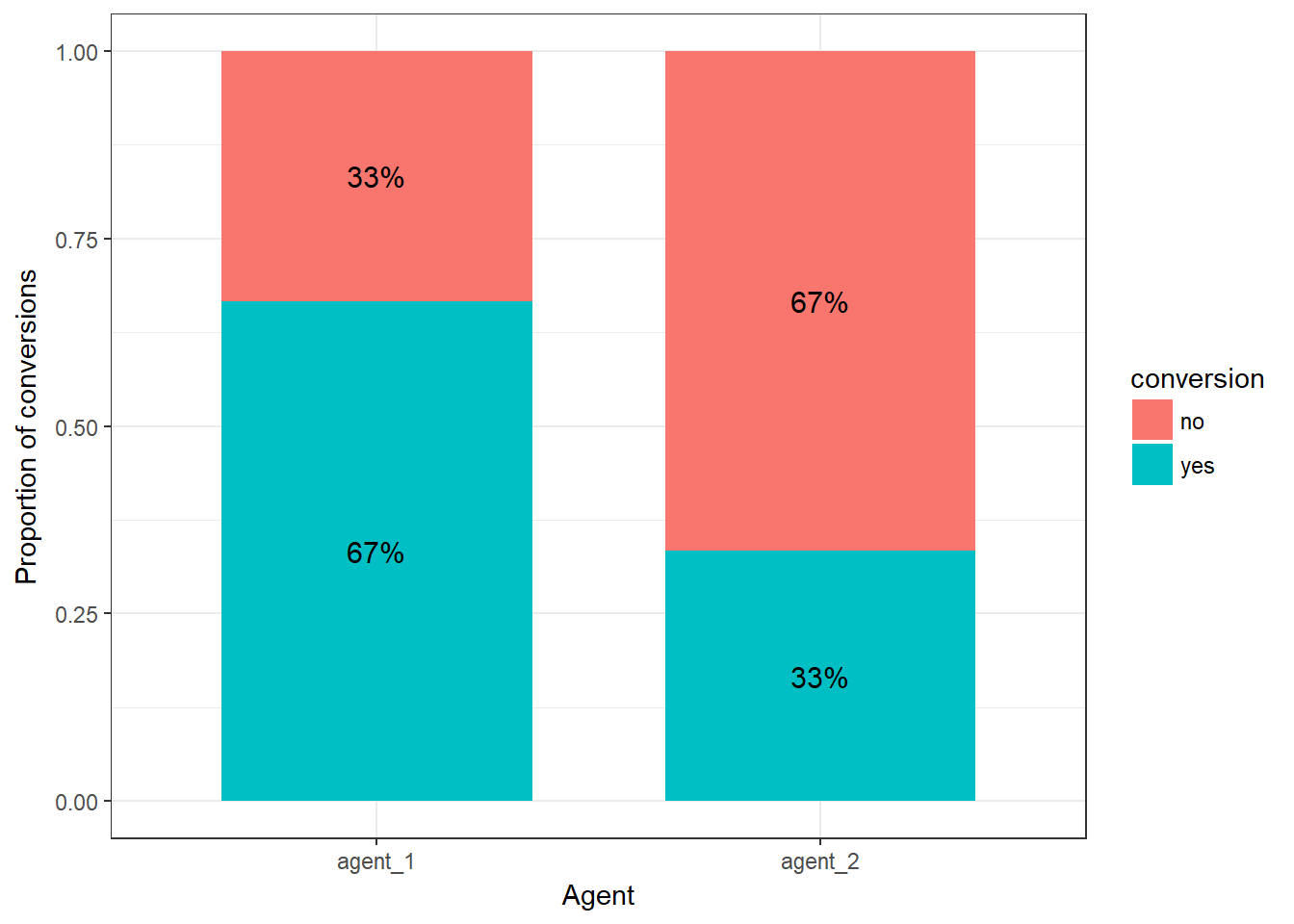
Figure 6.11: proportion of conversions per agent (stacked bar chart)
… or using the mosaicplot() function:
contigency_table <- table(call_center)
mosaicplot(contigency_table, main = "Proportion of conversions by agent")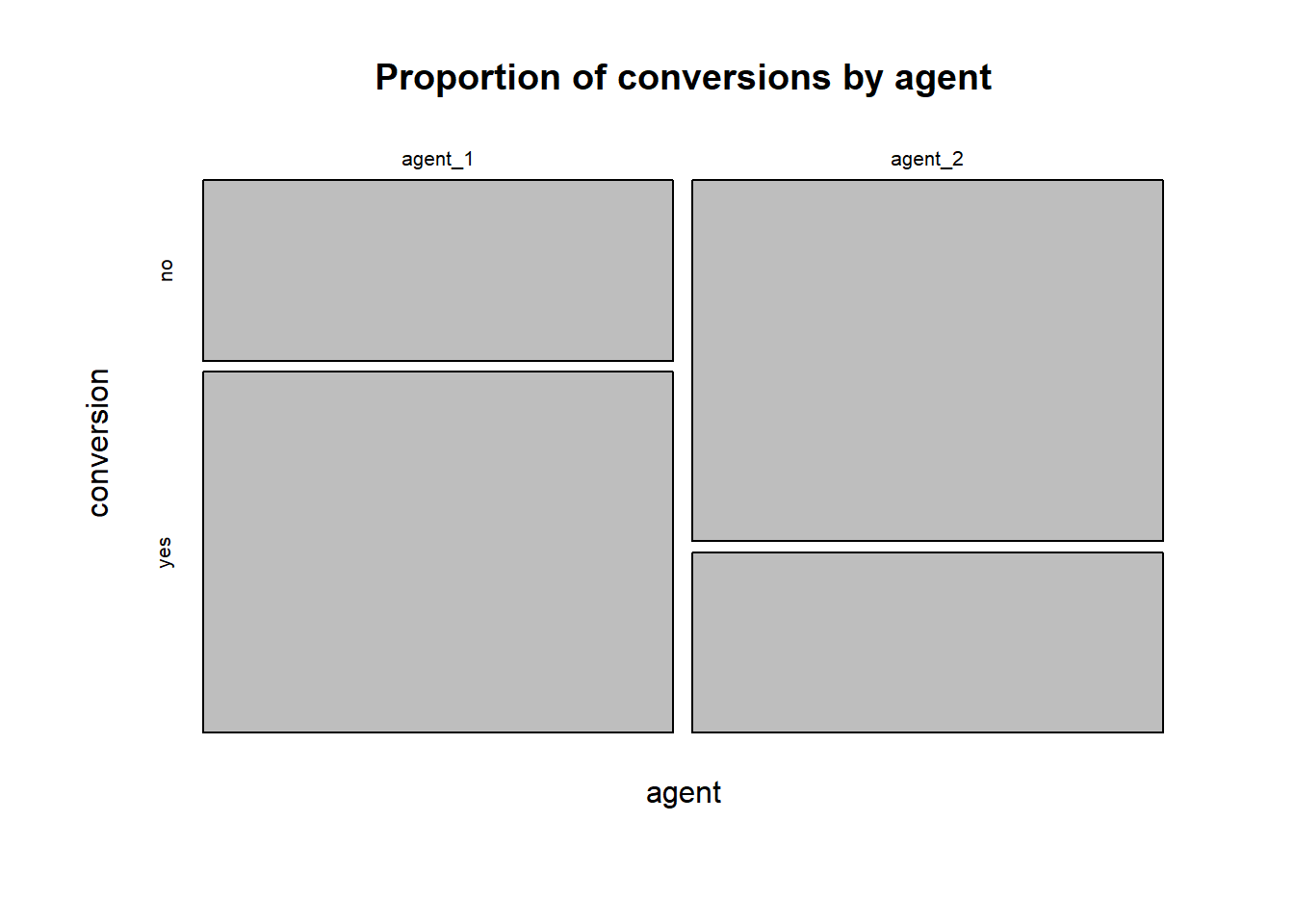
Figure 6.12: proportion of conversions per agent (mosaic plot)
Recall that we can use confidence intervals to determine the range of values that the true population parameter will take with a certain level of confidence based on the sample. Similar to the confidence interval for means, we can compute a confidence interval for proportions. The (1-α)% confidence interval for poportions is approximately
\[\begin{equation} \begin{split} CI = p\pm z_{1-\frac{\alpha}{2}}*\sqrt{\frac{p*(1-p)}{N}} \end{split} \tag{6.17} \end{equation}\]where \(\sqrt{p(1-p)}\) is the equvalent to the standard deviation in the formula for the confidence interval for means. Based on the equation, it is easy to compute the confidence intervals for the conversion rates of the call center agents:
n1 <- nrow(subset(call_center, agent == "agent_1")) #number of observations for agent 1
n2 <- nrow(subset(call_center, agent == "agent_2")) #number of observations for agent 1
n1_conv <- nrow(subset(call_center, agent == "agent_1" &
conversion == "yes")) #number of conversions for agent 1
n2_conv <- nrow(subset(call_center, agent == "agent_2" &
conversion == "yes")) #number of conversions for agent 2
p1 <- n1_conv/n1 #proportion of conversions for agent 1
p2 <- n2_conv/n2 #proportion of conversions for agent 2
error1 <- qnorm(0.975) * sqrt((p1 * (1 - p1))/n1)
ci_lower1 <- p1 - error1
ci_upper1 <- p1 + error1
ci_lower1## [1] 0.6133232ci_upper1## [1] 0.7200101error2 <- qnorm(0.975) * sqrt((p2 * (1 - p2))/n2)
ci_lower2 <- p2 - error2
ci_upper2 <- p2 + error2
ci_lower2## [1] 0.2799899ci_upper2## [1] 0.3866768Similar to testing for differences in means, we could also ask: Is agent 1 twice as likely as agent 2 to convert a customer? Or, to state it mathematically:
\(H_0: p_1=p_2\)
\(H_1: p_1\ne p_2\)
One approach to test this is based on confidence intervals to estimate the difference between two populations. We can compute an approximate confidence interval for the difference between the proportion of successes in group 1 and group 2, as:
\[\begin{equation} \begin{split} CI = p_1-p_2\pm z_{1-\frac{\alpha}{2}}*\sqrt{\frac{p_1*(1-p_1)}{n_1}+\frac{p_2*(1-p_2)}{n_2}} \end{split} \tag{6.17} \end{equation}\]If the confidence interval includes zero, then the data does not suggest a difference between the groups. Let’s compute the confidence interval for differences in the proportions by hand first:
ci_lower <- p1 - p2 - qnorm(0.975) * sqrt(p1 * (1 -
p1)/n1 + p2 * (1 - p2)/n2) #95% CI lower bound
ci_upper <- p1 - p2 + qnorm(0.975) * sqrt(p1 * (1 -
p1)/n1 + p2 * (1 - p2)/n2) #95% CI upper bound
ci_lower## [1] 0.2578943ci_upper## [1] 0.4087724Now we can see that the 95% confidence interval estimate of the difference between the proportion of conversions for agent 1 and the proportion of conversions for agent 2 is between 26% and 41%. This interval tells us the range of plausible values for the difference between the two population proportions. According to this interval, zero is not a plausible value for the difference (i.e., interval does not cross zero), so we reject the null hypothesis that the population proportions are the same.
Instead of computing the intervals by hand, we could also use the prop.test() function:
prop.test(x = c(n1_conv, n2_conv), n = c(n1, n2), conf.level = 0.95)##
## 2-sample test for equality of proportions with continuity
## correction
##
## data: c(n1_conv, n2_conv) out of c(n1, n2)
## X-squared = 65.34, df = 1, p-value = 0.0000000000000006303
## alternative hypothesis: two.sided
## 95 percent confidence interval:
## 0.2545610 0.4121057
## sample estimates:
## prop 1 prop 2
## 0.6666667 0.3333333Note that the prop.test() function uses a slightly different (more accurate) way to compute the confidence interval (Wilson’s score method is used). It is particularly a better approximation for smaller N. That’s why the confidence interval in the output slightly deviates from the manual computation above, which uses the Wald interval.
You can also see that the output from the prop.test() includes the results from a χ2 test for the equality of proportions (which will be discussed below) and the associated p-value. Since the p-value is less than 0.05, we reject the null hypothesis of equal probability. Thus, the reporting would be:
The test showed that the conversion rate for agent 1 was higher by 33%. This difference is significant χ (1) = 70, p < .05 (95% CI = [0.25,0.41]).
To calculate the required sample size when comparing porportions, the power.prop.test() function can be used. For example, we could ask how large our sample needs to be if we would like to compare two groups with probabilities of 10% and 15%, respectively using the conventional settings for α and β:
power.prop.test(p1 = 0.01, p2 = 0.15, sig.level = 0.05,
power = 0.8)##
## Two-sample comparison of proportions power calculation
##
## n = 57.75355
## p1 = 0.01
## p2 = 0.15
## sig.level = 0.05
## power = 0.8
## alternative = two.sided
##
## NOTE: n is number in *each* groupThe output tells us that we need 58 observations per group to detect a difference of the desired size.
6.4.2 Chi-square test
We came across the χ2 test in the previous section when we used it to test for the equality of proportions. Whenever you would like to investigate the relationship between two categorical variables, the χ2 test may be used to test whether the variables are independent of each other. It achieves this by comparing the expected number of observations in a group to the actual values. Consider the data set below, where each survey participant either owns an expensive car (coded as a 1) or doesn’t, and either is college educated (coded as a 1) or not (example based on N.K. Malhotra (2010): Marketing Research: An Applied Orientation). Let’s create the contingency table first:
cross_tab <- read.table("https://raw.githubusercontent.com/IMSMWU/Teaching/master/MRDA2017/cross_tab.dat",
sep = "\t", header = TRUE) #read data
cross_tab$College <- factor(cross_tab$College, levels = c(0:1),
labels = c("no", "yes")) #convert to factor
cross_tab$CarOwnership <- factor(cross_tab$CarOwnership,
levels = c(0:1), labels = c("no", "yes")) #convert to factor
cont_table <- table(cross_tab) #create contigency table
cont_table #view table## CarOwnership
## College no yes
## no 590 160
## yes 170 80To get a first impression regarding the association between the two variable, we compute the conditional relative frequencies and plot the observed shares by group:
cont_table_df <- as.data.frame(prop.table(table(cross_tab),1)) #conditional relative frequencies
cont_table_dfggplot(cont_table_df, aes(x = College, y = Freq, fill = CarOwnership)) + #plot data
geom_col(width = .7) + #position
geom_text(aes(label = paste0(round(Freq*100,0),"%")), position = position_stack(vjust = 0.5), size = 4) + #add percentages
ylab("Expensive car ownership (proportion)") + xlab("College degree") + # specify axis labels
theme_bw()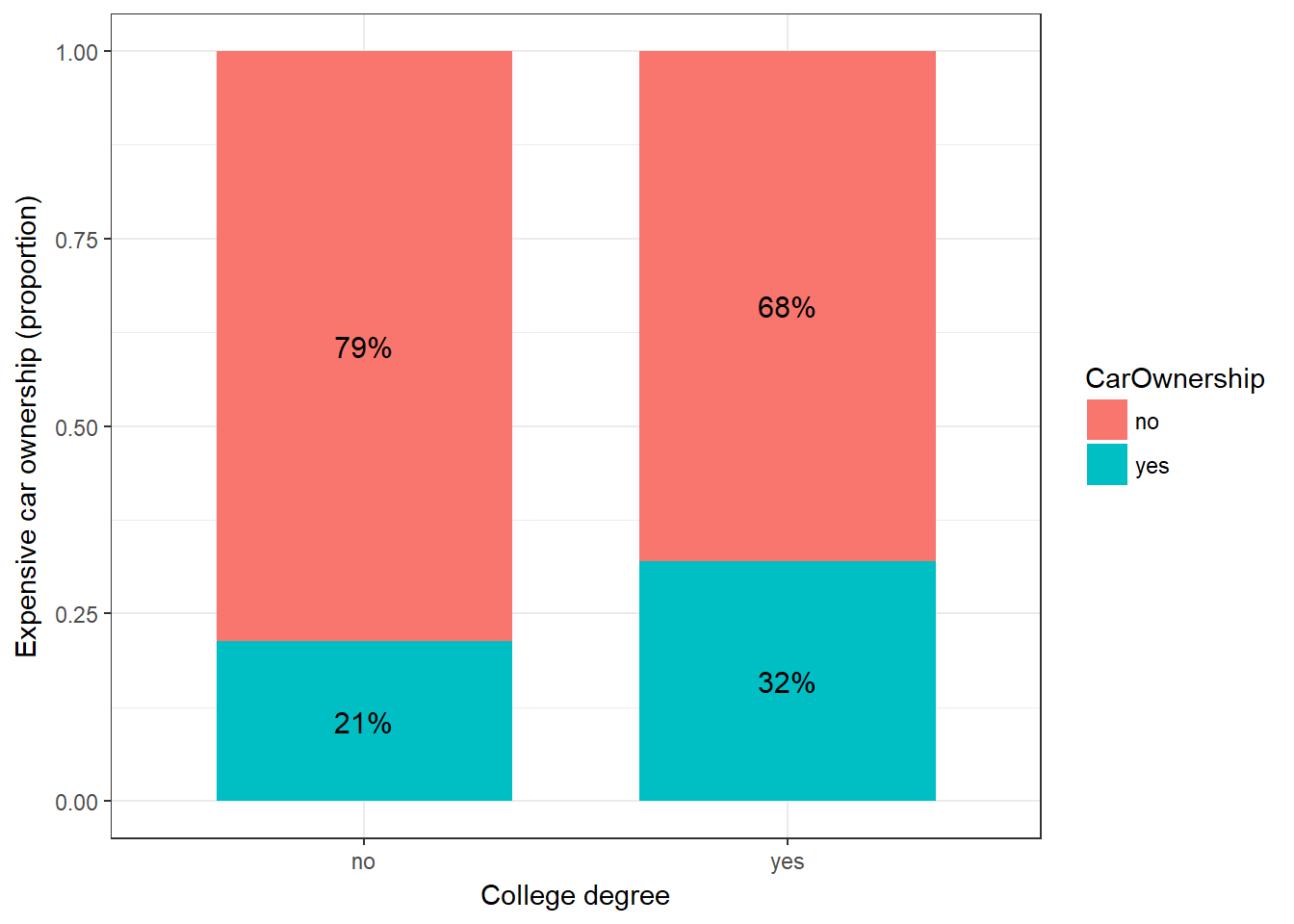
Figure 6.13: Expensive car ownership conditional on college education
Under the null hypothesis, the two variables are independent (i.e., there is no relationship). This means that the frequency in each field will be roughly proportional to the probability of an observation being in that category, calculated under the assumption that they are independent. The difference between that expected quantity and the actual quantity can be used to construct the test statistic. The test statistic is computed as follows:
\[\begin{equation} \begin{split} \chi^2=\sum_{i=1}^{J}\frac{(f_o-f_e)^2}{f_e} \end{split} \tag{6.18} \end{equation}\]where \(J\) is the number of cells in the contigency table, \(f_o\) are the observed cell frequencies and \(f_e\) are the expected cell frequencies. The larger the differences, the larger the test statistic and the smaller the p-value.
The observed cell frequencies can easily be seen from the contingency table:
obs_cell1 <- cont_table[1, 1]
obs_cell2 <- cont_table[1, 2]
obs_cell3 <- cont_table[2, 1]
obs_cell4 <- cont_table[2, 2]The expected cell frequencies can be calculated as follows:
\[\begin{equation} \begin{split} f_e=\frac{(n_r*n_c)}{n} \end{split} \tag{6.19} \end{equation}\]where \(n_r\) are the total observed frequencies per row, \(n_c\) are the total observed frequencies per column, and \(n\) is the total number of observations. Thus, the expected cell frequencies under the assumption of independence can be calculated as:
n <- nrow(cross_tab)
exp_cell1 <- (nrow(cross_tab[cross_tab$College == "no",
]) * nrow(cross_tab[cross_tab$CarOwnership == "no",
]))/n
exp_cell2 <- (nrow(cross_tab[cross_tab$College == "no",
]) * nrow(cross_tab[cross_tab$CarOwnership == "yes",
]))/n
exp_cell3 <- (nrow(cross_tab[cross_tab$College == "yes",
]) * nrow(cross_tab[cross_tab$CarOwnership == "no",
]))/n
exp_cell4 <- (nrow(cross_tab[cross_tab$College == "yes",
]) * nrow(cross_tab[cross_tab$CarOwnership == "yes",
]))/nTo sum up, these are the expected cell frequencies
data.frame(Car_no = rbind(exp_cell1, exp_cell2), Car_yes = rbind(exp_cell3,
exp_cell4), row.names = c("College_no", "College_yes"))## Car_no Car_yes
## College_no 570 190
## College_yes 180 60… and these are the observed cell frequencies
data.frame(Car_no = rbind(obs_cell1, obs_cell2), Car_yes = rbind(obs_cell3,
obs_cell4), row.names = c("College_no", "College_yes"))## Car_no Car_yes
## College_no 590 170
## College_yes 160 80To obtain the test statistic, we simply plug the values into the formula:
chisq_cal <- sum(((obs_cell1 - exp_cell1)^2/exp_cell1),
((obs_cell2 - exp_cell2)^2/exp_cell2), ((obs_cell3 -
exp_cell3)^2/exp_cell3), ((obs_cell4 - exp_cell4)^2/exp_cell4))
chisq_cal## [1] 11.69591The test statistic is \(\chi^2\) distributed. The chi-square distribution is a non-symmetric distribution. Actually, there are many different chi-square distributions, one for each degree of freedom as show in the following figure.
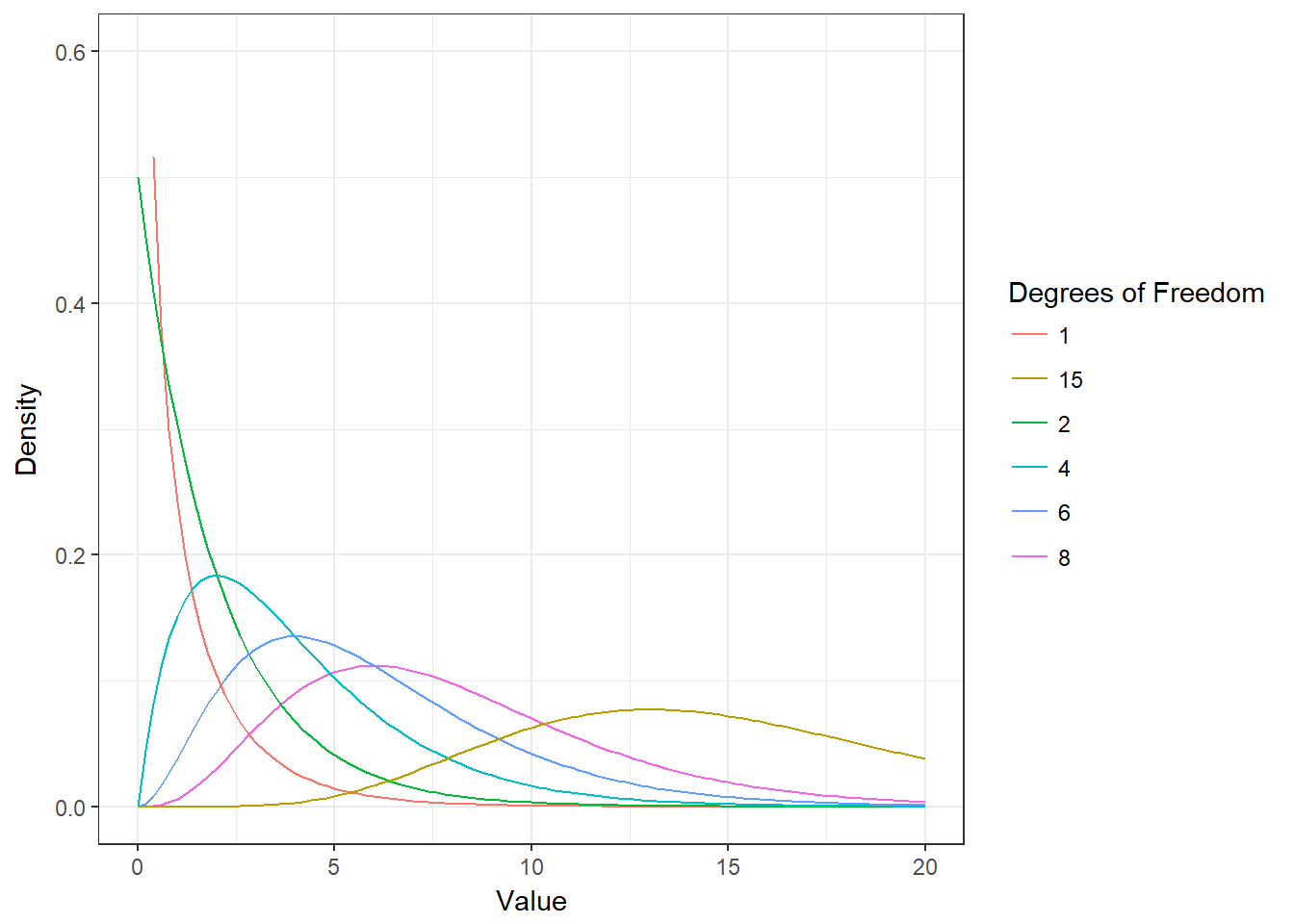
Figure 6.14: The chi-square distribution
You can see that as the degrees of freedom increase, the chi-square curve approaches a normal distribution. To find the critical value, we need to specify the corresponding degrees of freedom, given by:
\[\begin{equation} df=(r-1)*(c-1) \tag{6.20} \end{equation}\]where \(r\) is the number of rows and \(c\) is the number of columns in the contingency table. Recall that degrees of freedom are generally the number of values that can vary freely when calculating a statistic. In a 2 by 2 table as in our case, we have 2 variables (or two samples) with 2 levels and in each one we have 1 that vary freely. Hence, in our example the degrees of freedom can be calculated as:
df <- (nrow(cont_table) - 1) * (ncol(cont_table) -
1)
df## [1] 1Now, we can derive the critical value given the degrees of freedom and the level of confidence using the qchisq() function and test if the calculated test statistic is larger than the critical value:
chisq_crit <- qchisq(0.95, df)
chisq_crit## [1] 3.841459chisq_cal > chisq_crit## [1] TRUE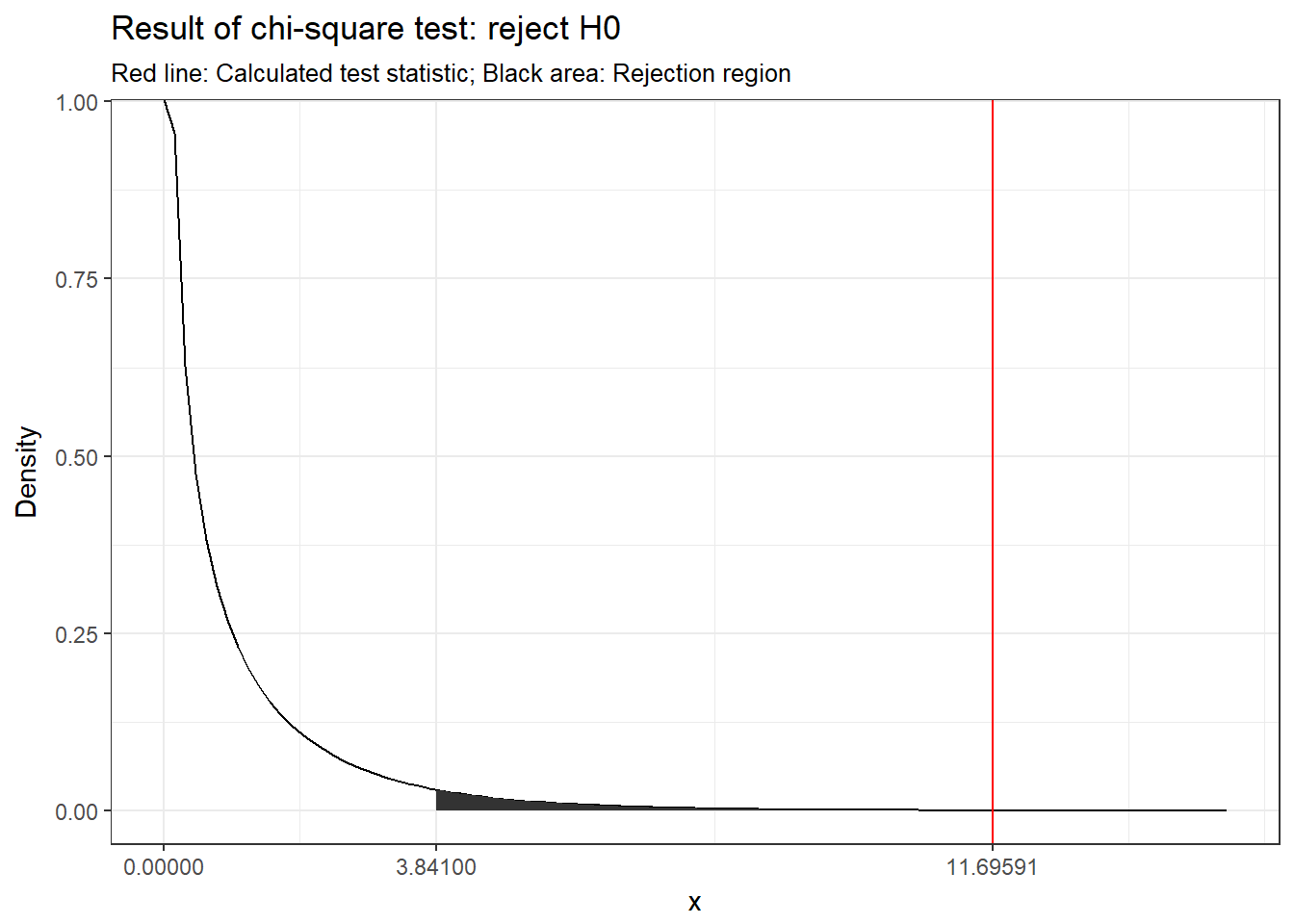
Figure 6.15: Visual depiction of the test result
We could also compute the p-value using the pchisq() function, which tells us the probability of the observed cell frequencies if the null hypothesis was true (i.e., there was no association):
p_val <- 1 - pchisq(chisq_cal, df)
p_val## [1] 0.0006263775The test statistic can also be calculated in R directly on the contigency table with the function chisq.test().
chisq.test(cont_table, correct = FALSE)##
## Pearson's Chi-squared test
##
## data: cont_table
## X-squared = 11.696, df = 1, p-value = 0.0006264Since the p-value is smaller than 0.05 (i.e., the calculated test statistic is larger than the critical value), we reject H0 that the two variables are independent.
Note that the test statistic is sensitive to the sample size. To see this, lets assume that we have a sample of 100 observations instead of 1000 observations:
chisq.test(cont_table/10, correct = FALSE)##
## Pearson's Chi-squared test
##
## data: cont_table/10
## X-squared = 1.1696, df = 1, p-value = 0.2795You can see that even though the proportions haven’t changed, the test is insignificant now. The following equation let’s you compute a measure of the effect size, which is insensitive to sample size:
\[\begin{equation} \begin{split} \phi=\sqrt{\frac{\chi^2}{n}} \end{split} \tag{6.19} \end{equation}\]The following guidelines are used to determine the magnitude of the effect size (Cohen, 1988):
- 0.1 (small effect)
- 0.3 (medium effect)
- 0.5 (large effect)
In our example, we can compute the effect sizes for the large and small samples as follows:
test_stat <- chisq.test(cont_table, correct = FALSE)$statistic
phi1 <- sqrt(test_stat/n)
test_stat <- chisq.test(cont_table/10, correct = FALSE)$statistic
phi2 <- sqrt(test_stat/(n/10))
phi1## X-squared
## 0.1081476phi2## X-squared
## 0.1081476You can see that the statistic is insensitive to the sample size.
Note that the Φ coefficient is appropriate for two dichotomous variables (resulting from a 2 x 2 table as above). If any your nominal variables has more than two categories, Cramér’s V should be used instead:
\[\begin{equation} \begin{split} V=\sqrt{\frac{\chi^2}{n*df_{min}}} \end{split} \tag{6.21} \end{equation}\]where \(df_{min}\) refers to the degrees of freedom associated with the variable that has fewer categories (e.g., if we have two nominal variables with 3 and 4 categories, \(df_{min}\) would be 3 - 1 = 2). The degrees of freedom need to be taken into account when judging the magnitude of the effect sizes (see e.g., here).
Note that the correct = FALSE argument above ensures that the test statistic is computed in the same way as we have done by hand above. By default, chisq.test() applies a correction to prevent overestimation of statistical significance for small data (called the Yates’ correction). The correction is implemented by subtracting the value 0.5 from the computed difference between the observed and expected cell counts in the numerator of the test statistic (see Equation (6.18)). This means that the calculated test statistic will be smaller (i.e., more conservative). Although the adjustment may go too far in some instances, you should generally rely on the adjusted results, which can be computed as follows:
chisq.test(cont_table)##
## Pearson's Chi-squared test with Yates' continuity correction
##
## data: cont_table
## X-squared = 11.118, df = 1, p-value = 0.0008547As you can see, the results don’t change much in our example, since the differences between the observed and expected cell frequencies are fairly large relative to the correction.
Caution is warranted when the cell counts in the contingency table are small. The usual rule of thumb is that all cell counts should be at least 5 (this may be a little too stringent though). When some cell counts are too small, you can use Fisher’s exact test using the fisher.test() function.
fisher.test(cont_table)##
## Fisher's Exact Test for Count Data
##
## data: cont_table
## p-value = 0.0008358
## alternative hypothesis: true odds ratio is not equal to 1
## 95 percent confidence interval:
## 1.243392 2.410325
## sample estimates:
## odds ratio
## 1.734336The Fisher test, while more conservative, also shows a significant difference between the proportions (p < 0.05). This is not surprising since the cell counts in our example are fairly large.